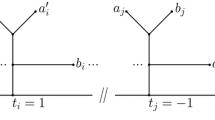
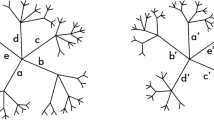
Abstract
A tree representation distance method, applied to any dissimilarity array, always gives a valued tree, even if the tree model is not appropriate. In the first part, we propose some criteria to evaluate the quality of the computed tree. Some of them are metric; their values depend on the edge’s lengths. The other ones only depend on the tree topology. In the second part, we calculate the average and the critical values of these criteria, according to parameters. Three models of distance are tested using simulations. On the one hand, the tree model, and on the other hand, euclidean distances, and boolean distances. In each case, we select at random distances fitting these models and add some noise. We show that the criteria values permit one to differentiate the tree model from the others. Finally, we analyze a distance between proteins and its tree representation that is valid according to the criteria values.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Bartélemy J.P. and Guenoche A.: Trees and Proximity Representations, J. Wiley (1991)
Gascuel 0. and Jean-Marie A.: Classification par arbre: la forme de l’arbre inféré dépend du schema algorithmique retenu, Actes des Septiémes Journées de la Société Francophone de Classification, SFC’99 (1999) 339–343
Guénoche A. and Préa P.: Counting and selecting at random phylogenetic topologies to compare reconstruction methods, Proceedings of the Conference of the International Federation of the Classifications Societies, IFCS’98, Short papers volume (1998), 242–245
Harding E.F.: The probabilities of rooted-tree shapes generated by random bifurcations, Advances in Applied Probabilities, 3 (1971) 44–77
Perriére and Gouy M.: www-Query: An on-line retrieval system for biological sequence banks. Biochimie, 78 (1996) 364–369
Pruzansky S., Tversky A. and Carroll J.D.: Spatial versus tree representations of proximity data, Psychometrika, 47, 1 (1982) 3–19
Saitou N. and Nei M.: The neighbor-joining method: a new method for reconstructing phylogenetic trees, Mol. Biol. Evol., 4 (1987) 406–425
Studier J.A. and Keppler K.J.: A note on the neighbor-joining method of Saitou and Nei, Mol. Biol. Evol., 5 (1988) 729–731.
Wu L.F., Ize B., Chanal A., Quentin Y. and Fichant G.: Bacterial Twin-arginine signal peptide-dependant protein translocation pathway: evolution and mechanism, J. Molecular Microbiology and Biotechnology (2000) (in press)
Wu L.F., Ize B., Chanal A., Quentin Y. and Fichant G.: Bacterial Twin-arginine signal peptide-dependant protein translocation pathway: evolution and mechanism, J. Molecular Microbiology and Biotechnology (2000) (in press)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2001 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Guénoche, A., Garreta, H. (2001). Can We Have Confidence in a Tree Representation?. In: Gascuel, O., Sagot, MF. (eds) Computational Biology. JOBIM 2000. Lecture Notes in Computer Science, vol 2066. Springer, Berlin, Heidelberg. https://doi.org/10.1007/3-540-45727-5_5
Download citation
DOI: https://doi.org/10.1007/3-540-45727-5_5
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-540-42242-6
Online ISBN: 978-3-540-45727-5
eBook Packages: Springer Book Archive