Abstract
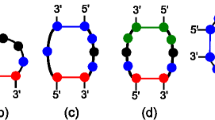
We view the folding of RNA-sequences as a map that assigns a pattern of base pairings to each sequence, known as secondary structure. These preimages can be constructed as random graphs (i.e. the neutral networks associated to the structure s).
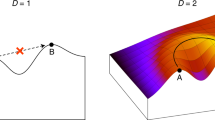
By interpreting the secondary structure as biological information we can formulate the so called Error Threshold of Shapes as an extension of Eigen's et al. concept of an error threshold in the single peak landscape [5]. Analogue to the approach of Derrida & Peliti [3] for a flat landscape we investigate the spatial distribution of the population on the neutral network.
On the one hand this model of a single shape landscape allows the derivation of analytical results, on the other hand the concept gives rise to study various scenarios by means of simulations, e.g. the interaction of two different networks [29]. It turns out that the intersection of two sets of compatible sequences (with respect to the pair of secondary structures) plays a key role in the search for “fitter” secondary structures.
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
L. W. Buss. The Evolution of Individuality. Princeton University Press, Princeton, 1987.
R. Dawkins. Replicator selection and the extended phenotype. Zeitschrift für Tierpsychologie, 47:61–76, 1978.
B. Derrida and L. Peliti. Evolution in a flat fitness landscape. Bull. Math. Biol., 53:355–382, 1991.
M. Eigen. Selforganization of matter and the evolution of biological macro-molecules. Die Naturwissenschaften, 10:465–523, 1971.
M. Eigen, J. McCaskill, and P. Schuster. The molecular Quasispecies. Adv. Chem. Phys., 75:149–263, 1989.
M. Eigen and P. Schuster. The Hypercycle: a principle of natural self-organization. Springer, Berlin, 1979 (ZBP:234.
W. Feller. An Introduction to Probability Theory and its Applications, volume I and II. John Wiley, New York, London, Sydney, 1966.
W. Fontana, T. Griesmacher, W. Schnabl, P. Stadler, and P. Schuster. Statistics of landscapes based on free energies, replication and degredation rate constants of RNA secondary structures. Monatshefte der Chemie, 122:795–819, 1991.
W. Fontana, D. A. M. Konings, P. F. Stadler, and P. Schuster. Statistics of RNA secondary structures. Biopolymers, 33:1389–1404, 1993.
W. Fontana, W. Schnabl, and P. Schuster. Physical aspects of evolutionary optimization and adaption. Physical Review A, 40(6):3301–3321, Sep. 1989.
W. Fontana, P. F. Stadler, E. G. Bornberg-Bauer, T. Griesmacher, I. L. Hofacker, M. Tacker, P. Tarazona, E. D. Weinberger, and P. Schuster. RNA folding and combinatory landscapes. Phys. Rev. E, 47(3):2083–2099, March 1993.
C. V. Forst, C. Reidys, and P. Schuster. Error thresholds, diffusion, and neutral networks. Artificial Life, 1995. in prep.
C. V. Forst, J. Weber, C. Reidys, and P. Schuster. Transitions and evolutive optimization in Multi Shape landscapes. in prep., 1995.
D. Gillespie. Exact stochastic simulation of coupled chemical reactions. J. Chem. Phys., 81:2340–2361, 1977.
I. L. Hofacker, W. Fontana, P. F. Stadler, S. Bonhoeffer, M. Tacker, and P. Schuster. Fast folding and comparison of RNA secondary structures. Monatshefte f. Chemie, 125(2):167–188, 1994.
M. Huynen, P. F. Stadler, and W. Fontana. Evolutionary dynamics of RNA and the neutral theory. Nature, 1994. submitted.
S. Karlin and H. M. Taylor. A first course in stochastic processes. Academic Press, second edition, 1975.
M. Kimura. Evolutionary rate at the molecular level. Nature, 217:624–626, 1968.
M. Kimura. The Neutral Theory of Molecular Evolution. Cambridge Univ. Press, Cambridge, UK, 1983.
D. R. Mills, R. L. Peterson, and S. Spiegelman. An extracellular darwinian experiment with a self-duplicating nucleic acid molecule. Proc. Nat. Acad. Sci., USA, 58:217–224, 1967.
M. Nowak and P. Schuster. Error tresholds of replication in finite populations, mutation frequencies and the onset of Muller's ratchet. Journal of theoretical Biology, 137:375–395, 1989.
C. Reidys, C. V. Forst, and P. Schuster. Replication on neutral networks of RNA secondary structures. Bull. Math. Biol., 1995. submitted.
C. Reidys, P. F. Stadler, and P. Schuster. Generic properties of combinatory maps and neutral networks of RNA secondary structures. Bulletin of Math. Biol., 1995. submitted.
P. Schuster, W. Fontana, P. F. Stadler, and I. L. Hofacker. From sequences to shapes and back: A case study in RNA secondary structures. Proc.Roy.Soc.(London)B, 255:279–284, 1994.
P. Schuster and P. F. Stadler. Landscapes: Complex optimization problems and biomolecular structures. Computers Chem., 18:295–324, 1994.
M. Tacker, W. Fontana, P. Stadler, and P. Schuster. Statistics of RNA melting kinetics. Eur. Biophys. J., 23(1):29–38, 1994.
M. Tacker, P. F. Stadler, E. G. Bornberg-Bauer, I. L. Hofacker, and P. Schuster. Robust Properties of RNA Secondary Structure Folding Algorithms. in preparation, 1993.
G. P. Wagner. What has survived of Darwin's theory? Evolutionary trends in plants, 4(2):71–73, 1990.
J. Weber, C. Reidys, and P. Schuster. Evolutionary optimization on neutral networks of RNA secondary structures. in preparation, 1995.
S. Wolfram. Mathematica: a system for doing mathematics by computer. Addison-Wesley, second edition, 1991.
S. Wright. The roles of mutation, inbreeding, crossbreeeding and selection in evolution. In D. F. Jones, editor, int. Proceedings of the Sixth International Congress on Genetics, volume 1, pages 356–366, 1932.
S. Wright. Random drift and the shifting balance theory of evolution. In K. Kojima, editor, Mathematical Topics in Population Genetics, pages 1–31. Springer Verlag, Berlin, 1970.
Author information
Authors and Affiliations
Corresponding author
Editor information
Rights and permissions
Copyright information
© 1995 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Forst, C.V., Reidys, C., Weber, J. (1995). Evolutionary dynamics and optimization. In: Morán, F., Moreno, A., Merelo, J.J., Chacón, P. (eds) Advances in Artificial Life. ECAL 1995. Lecture Notes in Computer Science, vol 929. Springer, Berlin, Heidelberg. https://doi.org/10.1007/3-540-59496-5_294
Download citation
DOI: https://doi.org/10.1007/3-540-59496-5_294
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-540-59496-3
Online ISBN: 978-3-540-49286-3
eBook Packages: Springer Book Archive