Abstract
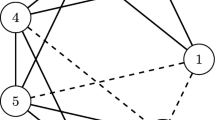
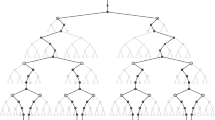
The discretizable molecular distance geometry problem (DMDGP) is a subclass of the MDGP, where instances can be solved using a discrete algorithm called branch-and-prune (BP). We present an initial study showing that the BP algorithm can be used differently from its original form, where the initial atoms are fixed and the branches of the BP tree are generated until the last atom is reached. Particularly, we show that the use of multiple BP trees may explore the search space faster than the original BP.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., Bourne, P.E.: The protein data bank. Nucleic Acids Res. 28, 235–242 (2000)
Crippen, G., Havel, T.: Distance Geometry and Molecular Conformation. Wiley, New York (1988)
Dong, Q., Wu, Z.: A linear-time algorithm for solving the molecular distance geometry problem with exact interatomic distances. J. Global Optim. 22, 365–375 (2002)
Lavor, C., Liberti, L., Maculan, N.: Molecular distance geometry problem. Encyclopedia of Optimization, pp. 2305–2311, 2nd edn. Springer, New York (2009)
Lavor, C., Liberti, L., Maculan, N., Mucherino, A.: The discretizable molecular distance geometry problem. Comput. Optim. Appl. 52, 115–146 (2012)
Lavor, C., Liberti, L., Maculan, N., Mucherino, A.: Recent advances on the discretizable molecular distance geometry problem. Eur. J. Oper. Res. 219, 698–706 (2012)
Liberti, L., Lavor, C., Maculan, N.: A branch-and-prune algorithm for the molecular distance geometry problem. Int. Trans. Oper. Res. 15, 1–17 (2008)
Liberti, L., Lavor, C., Mucherino, A., Maculan, N.: Molecular distance geometry methods: from continuous to discrete. Int. Trans. Oper. Res. 18, 33–51 (2010)
Mucherino, A., Lavor, C., Liberti, L., Talbi, E.-G.: A parallel version of the branch and prune algorithm for the molecular distance geometry problem. In: IEEE Conference Proceedings, ACS/IEEE International Conference on Computer Systems and Applications (AICCSA10), pp. 1–6. Hammamet, Tunisia (2010)
Nucci, P.: Heuristicas para o problema molecular de geometria de distancias aplicado a proteinas. Undergraduate Dissertation, Fluminense Federal University
Saxe, J.B.: Embeddability of weighted graphs in k-space is strongly NP-hard. In: Proceedings of 17th Allerton Conference in Communications, Control and Computing, pp. 480–489. Monticello, IL (1979)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer Science+Business Media New York
About this chapter
Cite this chapter
Nucci, P., Nogueira, L.T., Lavor, C. (2013). Solving the Discretizable Molecular Distance Geometry Problem by Multiple Realization Trees. In: Mucherino, A., Lavor, C., Liberti, L., Maculan, N. (eds) Distance Geometry. Springer, New York, NY. https://doi.org/10.1007/978-1-4614-5128-0_9
Download citation
DOI: https://doi.org/10.1007/978-1-4614-5128-0_9
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4614-5127-3
Online ISBN: 978-1-4614-5128-0
eBook Packages: Mathematics and StatisticsMathematics and Statistics (R0)