Abstract
The quantification of muscle mass is important in clinical populations with chronic paralysis, cachexia, and sarcopenia. This is especially true when testing interventions which are designed to maintain or improve muscle mass. The purpose of this paper is to report on an automated method of MRI-based thigh muscle segmentation framework that minimizes longitudinal deviation by using femur segmentation as a reference in a two-phase registration. Imaging data from seven patients with severe multiple sclerosis who had undergone MRI scans at multiple time points were used to develop and validate our method. The proposed framework results in robust, automated co-registration between baseline and follow up scans, and generates a reliable thigh muscle mask that excludes intramuscular fat.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
1 Introduction
Multiple sclerosis (MS) is an inflammatory disease of the central nervous system that can affect electrical conduction in axons in the brain, spinal cord, and optic nerves [3]. The clinical manifestations are partly driven by the location of focal inflammatory lesions and include paralysis/paresis, spasticity, fatigue, cognitive impairment, and sphincter dysfunction.
Regular exercise may benefit patients with MS, primarily by ameliorating deconditioning associated with disability. However, disability associated with advanced disease (Expanded Disability Status Scale (EDSS) of 7.0 or more) impedes regular exercise in this patient cohort [11, 12]. Previous exercise interventions in patients with advanced MS have been short-term and largely limited to arm exercise. Recently, studies have begun to investigate the potential of electrical stimulation and other treatments to maintain leg muscle mass. To assess the efficacy of these treatments in MS, a precise and accurate quantification of limb muscle mass change is critical.
In this study, we assessed the change in longitudinal thigh muscle volume in patients with advanced MS undergoing an exercise regime based on a modified version of NMES (Neuromuscular Electrical Stimulation) cycling [5, 6]. This work describes an automated segmentation pipeline to rapidly quantitate lean thigh muscle volume, and accurately estimate change over time.
Previously described methods for segmenting thigh muscle volume from MR images have utilised semi-automated approaches, including threshold algorithms and model matching techniques [2, 9]. More recently, semantic segmentation has become a popular approach for solving computer vision problems; and attempts have been made to segment muscle masks from MR images with deep learning based algorithms [1]. However, these models normally do not take the intramuscular fat of subject into account. Additionally, longitudinal changes in thigh muscle ROIs (regions of interest) have been seldom studied, and most published methods [4, 10, 13] employ cross-sectional segmentation without indicating the method of alignment of ROIs between baseline and follow-up images.
Hence, we propose an automated segmentation framework to rapidly and accurately quantitate longitudinal change in thigh muscle volume from MR images. The framework uses a novel two-phase registration method to strictly align the relevant ROIs between baseline and follow-up; and has been evaluated in 7 subjects. Our results indicate that the proposed method achieves a consistent geometric alignment in longitudinal MR thigh images in terms of Complex Wavelet Structural Similarity Image Metric (CW-SSIM).
2 Method
The proposed automated longitudinal thigh muscle segmentation framework is shown in Fig. 1. Any number of 3D T1-weighted MR images of both left and right thighs are used as the input to generate the corresponding muscle segmentation masks. To facilitate comparative analysis, region of interest (ROI) is defined by the analyst based on the anatomical landmarks and extends from the axial slice immediately superior to the patella to the inferior border of the gluteus maximus muscle. The pipeline only requires the analyst to select these two ROI endpoints on the baseline scan.
To maintain longitudinal coherence, the femur, which remains morphologically stable over time, is segmented at each time point. A two-phase process is used to co-register follow-up to baseline images and derive follow up ROIs in the baseline space. Inhomogeneity correction, thresholding and morphological processing are then applied to generate the final thigh muscle masks. The details is described as follow.
2.1 Cropping
The input data for each patient/scan contains both thighs, and their relative position is slightly different at each time point. Direct co-registration between baseline and follow-up will lead to an inaccurate rigid alignment result. In our pipeline, thigh muscle analysis is performed separately for left and right thighs. To achieve this, as shown in Fig. 2(b), each 3D image is firstly projected to the axial plane by averaging the voxels along the superior-inferior direction, and then further averaged to L-R line to obtain the mean intensity profile as shown in Fig. 2(c). The 3D MR thigh image is divided into left and right thigh (black line, Fig. 2(c)) in the sagittal plane, located automatically by the interpeak nadir in the associated intensity plot.
2.2 Two-Phase Registration
External deformation of muscle during scanning, or longitudinal change in muscle morphology hampers geometric alignment and accurate co-registration of baseline and follow up 3D thigh images. This is a critical step when calculating volumetric change of a ROI over time. We use a modified form of FLIRT [7] for the registration in our framework. FLIRT was originally designed for longitudinal brain registration, and is based on determining the transformation (\(T^*\)) that minimizes the intensity-based cost function:
where BL and FU represent for baseline (reference) and follow-up respectively, T(FU) represents for the transformed FU, \(S_T\) is the set of all affine transformations and C(X, Y) is the cost function. Here we use the correlation ratio (CR) [8] as the cost function:
When applied in this study, the default registration uses the whole thigh as the reference target. Unlike soft tissues, changes in bone morphology are negligible over the observation period; we therefore elect to define the femur as the reference target and propose a two-phase registration process: in the first phase, the transformation \(T_{femur}\), is derived and “BL” and “FU” in Eqs. (1) and (2) are replaced with “\(BL_{femur}\)” and “\(FU_{femur}\)” respectively. \(T_{femur}\) is then applied to FU to obtain the \(FU_{reg}\). Since we snip the ROI of the thigh along inferior-superior direction on the baseline image, the same measurement area along the thigh at follow-up is obtained by aligning the \(BL_{ROI}\) to \(FU_{reg}\).
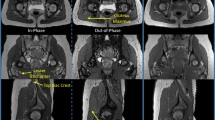
Figure 3 demonstrates scenarios where the use of the whole thigh as the registration reference fails to align the follow-ups with the baseline due to: (1) morphological change of muscle and fat (Fig. 3(a)); and (2) absence of distal thigh in the image (Fig. 3(b)).
2.3 Inhomogeneity Correction and Normalization
Inherent intensity inhomogeneity in MR images requires correction before a fixed threshold is applied for the segmentation of muscle from ROIs. In our pipeline, N4 [16], an improved version of the N3 framework [15], was used. N4 achieves the result by feeding the input image iteratively into a smoothing operator which contains a B-spline approximator.
2.4 Morphological Processing
After intensity inhomogeneity-correction and normalization, a fixed intensity threshold is used to segment the thigh muscle from surrounding tissues and differentiate the muscle from the intramuscular fat. To eliminate the noisy “ring” between the outer boundary of thigh and the background, and also the border between the femur and the thigh muscle, we perform following morphological process: first performing an opening of the threshold mask then take its intersection with the original mask. The demonstration of intermediate steps is shown in Fig. 4.
3 Experimental Results
Seven participants (five females and two males) with progressive MS (median age: \(55\pm 6\) years old, EDSS: \(7.3\pm 0.6\), patients with this level of EDSS lose majority of their mobility functions) were recruited for this study from a multidisciplinary MS clinic. Participants were asked to maintain with their usual activity and exercise routines in the first 12 weeks (control period) of the study; they undertook NMES leg cycling exercises three times per week during the second 12 week study epoch. In order to obtain consistent and reliable femur imaging, patients with following scenarios were not included in the analysis: (a) inability to lie in a standard position, (b) hip replacement and (c) severe spasticity.
MRI scans (both thighs) were acquired at the time of enrolment (baseline; week 0), following the 12 week control period, and post 12 weeks of NMES training (week 24). Additionally, long-term data was acquired from two patients. In summary, 21 MRI exams (7 baseline, 7 mid-study and 7 post-NMES) were acquired on a GE Discovery MR750 Scanner with 32-channel torso coil. All subjects were scanned with a 3DT1 sequence (IRFSPGR, TE \(=\) 2.7 ms, TR \(=\) 6.5 ms, acquisition metrix \(=\) 480 \(\times \) 480, Slice thickness \(=\) 1 mm).
For qualitative evaluation of the co-registration between baseline and follow-ups, Complex Wavelet Structural Similarity Image Metric (CW-SSIM) was computed as previously described [14]. Based on CW-SSIM score, our method yielded a higher similarity between baseline femur and femur on the co-registered follow-ups as shown in Table 1.
4 Conclusion
In this paper, we propose an automated longitudinal thigh muscle segmentation framework to calculate muscle volume change over time to assess the treatment effect. The technique can be potentially modified to sample upper arm musculature, and is ideally suited to applications requiring calculation of longitudinal changes in lean muscle mass. Femur-based co-registration minimised registration error and resulted in improved baseline to follow up image alignment, as measured by CW-SSIM. Low longitudinal measurement error and automation suggest that our technique will be suited to future inclusion in large clinical trials, both in the physical therapy and pharmacotherapy domains.
References
Ahmad, E., Goyal, M., McPhee, J.S., Degens, H., Yap, M.H.: Semantic segmentation of human thigh quadriceps muscle in magnetic resonance images. arXiv preprint arXiv:1801.00415 (2018)
Ahmad, E., Yap, M.H., Degens, H., McPhee, J.S.: Atlas-registration based image segmentation of MRI human thigh muscles in 3D space. In: Medical Imaging 2014: Image Perception, Observer Performance, and Technology Assessment, vol. 9037, p. 90371L. International Society for Optics and Photonics (2014)
Crayton, H.J., Rossman, H.S.: Managing the symptoms of multiple sclerosis: a multimodal approach. Clin. Ther. 28(4), 445–460 (2006)
Ema, R., Wakahara, T., Yanaka, T., Kanehisa, H., Kawakami, Y.: Unique muscularity in cyclists’ thigh and trunk: a cross-sectional and longitudinal study. Scand. J. Med. Sci. Sport. 26(7), 782–793 (2016)
Fornusek, C., Hoang, P., Barnett, M., Oliver, R., Burns, J.: Effect of neuromuscular electrical stimulation cycling on thigh muscle mass and strength in persons with advanced multiple sclerosis. J. Neurol. Sci. 381, 248 (2017)
Fornusek, C., Davis, G.M., Sinclair, P.J., Milthorpe, B.: Development of an isokinetic functional electrical stimulation cycle ergometer. Neuromodulation Technol. Neural Interface 7(1), 56–64 (2004)
Jenkinson, M., Bannister, P., Brady, M., Smith, S.: Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage 17(2), 825–841 (2002)
Jenkinson, M., Smith, S.: A global optimisation method for robust affine registration of brain images. Med. Image Anal. 5(2), 143–156 (2001)
Kemnitz, J., et al.: Validation of a 3D thigh muscle and adipose tissue segmentation method using statistical shape models. Osteoarthr. Cartil. 26, S457–S458 (2018)
Kemnitz, J., Wirth, W., Eckstein, F., Culvenor, A.: Thigh muscle and adipose tissue changes during symptomatic and radiographic knee osteoarthritis progression–data from the osteoarthritis initiative. Osteoarthr. Cartil. 26, S406 (2018)
Kurtzke, J.F.: Rating neurologic impairment in multiple sclerosis: an expanded disability status scale (EDSS). Neurology 33(11), 1444 (1983)
Meyer-Moock, S., Feng, Y.S., Maeurer, M., Dippel, F.W., Kohlmann, T.: Systematic literature review and validity evaluation of the expanded disability status scale (EDSS) and the multiple sclerosis functional composite (MSFC) in patients with multiple sclerosis. BMC Neurol. 14(1), 58 (2014)
Ruhdorfer, A., Wirth, W., Eckstein, F.: Longitudinal change in thigh muscle strength prior to and concurrent with minimum clinically important worsening or improvement in knee function: data from the osteoarthritis initiative. Arthritis Rheumatol. 68(4), 826–836 (2016)
Sampat, M.P., Wang, Z., Gupta, S., Bovik, A.C., Markey, M.K.: Complex wavelet structural similarity: a new image similarity index. IEEE Trans. Image Process. 18(11), 2385–2401 (2009)
Tustison, N., Gee, J.: N4ITK: Nick’s N3 ITK implementation for MRI bias field correction. Insight J. 9 (2009)
Tustison, N.J., et al.: N4ITK: improved N3 bias correction. IEEE Trans. Med. Imaging 29(6), 1310–1320 (2010)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Tang, Z. et al. (2018). Automatic Segmentation of Thigh Muscle in Longitudinal 3D T1-Weighted Magnetic Resonance (MR) Images. In: Melbourne, A., et al. Data Driven Treatment Response Assessment and Preterm, Perinatal, and Paediatric Image Analysis. PIPPI DATRA 2018 2018. Lecture Notes in Computer Science(), vol 11076. Springer, Cham. https://doi.org/10.1007/978-3-030-00807-9_2
Download citation
DOI: https://doi.org/10.1007/978-3-030-00807-9_2
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00806-2
Online ISBN: 978-3-030-00807-9
eBook Packages: Computer ScienceComputer Science (R0)