Abstract
Improved outcome in patients with ischemic stroke is achieved through acute diagnosis and early restoration of cerebral flow in appropriate patients. Diffusion-weighted MR imaging (DWI) plays a central role in these efforts by enabling rapid early localization and quantification of ischemic lesions. Automated detection and quantification can potentially accelerate diagnosis, improve treatment safety and efficacy and reduce costs. However, the manual quantification of acute ischemic stroke volumes for algorithm training is time consuming and imprecise. We present YNet as a novel fully-automated deep learning algorithm for detection and volumetric segmentation and quantification of acute cerebral ischemic lesions from DWI. The algorithm is a semi-supervised multi-tasking deep neural network architecture we developed that enables the combination of both weak labels derived from radiology report classification and manually delineated pixel level training data. The model is trained on a very large dataset of 10000 studies, achieves detection sensitivity 0.981, detection specificity 0.980 and segmentation Dice score 0.623 on a heterogeneous test set.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Acute ischemic stroke remains a leading cause of death and disability. However, a number of recent multicenter trials have demonstrated significantly improved neurological outcomes when the latest generation of thrombectomy devices are used to restore cerebral blood flow within 6 h of symptom onset [1, 5]. Extension of the time window for successful treatment out to 24 h has been more recently demonstrated [6]. In such trials, proper patient selection with various neuroimaging modalities, including CT, catheter angiography, and MRI, has been essential for treatment success. With MRI, diffusion-weighted imaging (DWI) sequences provide sensitive detection and localization of even the tiniest ischemic brain lesions within minutes of vessel occlusion. From DWI images, the total amount of ischemic tissue can also be measured. While critical for safe patient selection and for prediction of long term disability, such volume measurements are challenging. Automated methods for DWI lesion detection and measurement have great potential for improving stroke treatment by reducing the time necessary to administer therapy to appropriately selected patients. Though several semi-automated approaches have been proposed in the literature, including adaptive thresholding [8], watershed [12] and fuzzy clustering [13], fully automated solutions are preferable to reduce time-consuming and error-prone steps in the clinical workflow. However, a key bottleneck in building fully-automated solutions is the segmentation models require voxel-level image annotation of each image which is expensive and time-consuming. For example, Chen et al. introduced a fully-automated deep learning segmentation algorithm for DWI ischemic lesions based on 2D convolutional neural networks (CNN) which achieved good results of dice score of 0.67 and sensitivity of 0.94, but required training on 741 manually segmented DWI images. Binary classification of diagnostic radiology reports for brain MRI studies as DWI-positive or DWI-negative can be used as a source of weak image-level labels and are far easier to generate than detailed pixel-level annotations. In this work, we introduce a semi-supervised multi-tasking deep learning CNN model trained on explicit classification and segmentation tasks. The model learns to segment and to classify from a heterogeneous set of annotated and weakly labeled images.
2 Data
Data for this project was collected from Massachusetts General Hospital and Brigham Women Hospital. All data were collected retrospectively. Ethical approval was granted by the institutional review board. Data included MR studies and the official diagnostic reports.
2.1 Weak Labels
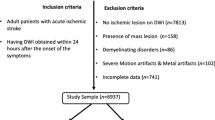
From a database containing all radiology reports over a ten year period (2007–2017), parsing of the Impression section text of Brain MRI study reports was performed to create a balanced set of 5000 negative studies and 5000 positive studies for acute ischemia. Parsing methods included keyword and sentence matching and creation of a simple text classifier based on N-grams and Support Vector Machines (SVM). Initial classification results were manually validated and corrected by a trained radiologist (B.B). Studies with keyword matches for terms indicating the presence of post-surgical findings or brain tumor pathology were excluded to reduce the effect of confounding image patterns from non-ischemic disease. The resultant scans were obtained from a variety of scanner manufacturers, magnet strengths, acquisition parameters and patient demographics. Patient age was limited to 20–80 years.
2.2 Manual Segmentation
Out of the 5000 positively labeled studies, 500 were manually segmented by two trained radiologists (B.B., R.R.) using the software Osirix version 9.0, by drawing contours of acute ischemic lesions on DWI and ADC series slices. In total, 1423 slices were manually segmented. The average manual segmentation time per volume was 10 min.
2.3 Imaging Data
The majority of ischemic lesion segmentation algorithms in the literature are based on DWI [2, 12, 13]. In this study, in order to mitigate shine-through effect from non-acute lesions, we incorporated both DWI and derived Apparent Diffusion Coefficient (ADC), following clinical practice. DWI and ADC images were acquired using 2D multi-slice MR acquisition modalities with variation in acquisition parameters (number of pixels, number of slices, pixels size, slice thickness, repetition time, echo time) related to different manufacturer platforms. Images were re-sampled in three dimensions on a grid of \(256\times 256 \times 32\) with resolution 1.2 mm \(\times \) 1.2 mm \(\times \) 9 mm. To compensate for the large variation in image intensity across studies, a normalization algorithm was applied independently to DWI and ADC image intensities. The algorithm detects the peak corresponding to white matter in the image histogram and scales the image to assign an average pixel value of 1.0 in the white matter.
3 Model
We developed YNet, an extension of the UNet architecture [3, 11], successfully employed by other authors for semantic segmentation of MR images [9], for our semi-supervised learning. The model operates on multichannel (DWI and ADC) image data at full resolution \((256\times 256 \times 32 \times 2)\).
3.1 Semi-Supervised Learning
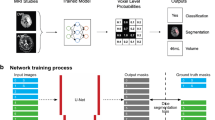
Figure 1 shows the architecture of the proposed YNet. The network is composed of three branches: encoding, decoding and classification. The encoding branch is composed of a cascade of convolutional blocks, each comprising two convolutional layers followed by max-pooling, batch normalization and activation. The decoding branch is composed of deconvolutional blocks, each comprising an up-sampling layer followed by two convolutional layers, batch normalization and activation. The classification branch, connected to the output of the encoder, is composed of two fully connected layers. The model loss is the sum of two terms: the negative Bernoulli log-likelihood associated to the output of the classification branch and the negative Dice score which measures the similarity of the output of the decoder branch to the manual segmentation. The Dice score previously used in [2, 9] as segmentation score, was modified as follows:
where \(x_i\) is the output of the decoder at voxel i; \(y_i\) is the manual segmentation at voxel i; \(\epsilon \) is a small number (set to 1.0 in experiments) for numerical stability; and \(m \in \{0,1\}\) is a binary mask variable that enables end-to-end training with mixed image-level and pixel-level annotations. The variable is set to 1 for positive manually segmented and negative cases, to 0 for positive non-segmented cases. This has the effect of disabling gradient propagation in the decoder when the pixels-level labeling is missing. From the perspective of image segmentation, the image-level labels provide semi-supervision. From the perspective of classification, the pixel-level labels act as a supervised focusing mechanism. A similar idea is applied in [10] to the semantic annotation of natural images. We implemented the model in Keras version 2.0 with TensorFlow back-end version 1.3.
4 Experiments and Results
The YNet model was compared with two baseline models: a convolutional neural network (CNN) for classification and a 3D UNet for segmentation. The CNN model for classification consisted of N convolutional layers. \(L_2\) regularization, dropout, and learning rates were explored through a distributed random search for 20 epochs. Configurations that performed well were then trained until convergence. After exploring a variety of architectures and hyperparameter configurations, the most performant model was selected based on the validation set accuracy. It consisted of six convolutional layers beginning with 36 filters and growing by 12 additional filters per layer. Test set sensitivity was observed to be 0.979 and specificity 0.958.
Experiments were performed using a 2D UNet to optimize the segmentation architecture. In order to select the optimal size of the receptive field, a 2D UNet with four encoding blocks and three decoding blocks was trained at three levels of resolution on all manually segmented slices: \(64\times 64 \times 32\), \(128\times 128 \times 32\) and \(256\times 256 \times 32\). The networks produced Dice coefficients of 0.527, 0.532, 0.561 respectively. The largest receptive field was selected and used in subsequent experiments.
The effect of various pre-processing techniques on segmentation performance was evaluated. Magnetic field inhomogeneity was corrected using the N4 Bias field correction software [14] and all images were rigidly registered to a reference DWI-ADC image pair. Bias field correction was found to degrade the segmentation performance. Image registration was found to slightly accelerate convergence but did not produce any significant improvement in segmentation performance. Bias field correction and image registration were therefore not used in subsequent experiments. In experiments with 2D segmentation, perturbation of the image intensity using random scale and offset was found to improve performance on the validation set. In subsequent experiments, the dataset was augmented 5-folds by multiplying the image intensity by \(\mathcal {N}(1,0.2)\) and by adding \(\mathcal {N}(0,0.2)\).
The final 3D Unet had a receptive field of \(256 \times 256 \times 32\) pixels with two input channels (DWI and ADC), five encoder blocks and four decoder blocks. Convolutional layers used \(3 \times 3 \times 3\) kernels and MaxPooling layers had pool size of \(2 \times 2 \times 2\). Dropout of 0.2 was used in the bottom layers of the encoding and decoding branches for regularization.
The YNet model architecture was identical to the 3D UNet architecture, with the addition of two dense layers for classification. The dense layers had size \(262144 \times 16\) and \(16 \times 1\) respectively. A dropout layer with dropout rate 0.5 was placed between the two dense layers for regularization.
In experiments, it was found that the performance of both the 3D UNet and the 3D YNet improved when pre-training the models in 2D. A 2D model with the same architecture and parameters as the 3D UNet was first trained on the set of all manually segmented 2D slices. The parameters were then transfered to the 3D UNet and YNet models, as depicted in Fig. 2, by padding the 2D kernels with random numbers generated according to the xavier initialization method [4]. During 2D pre-training, batch size was set to 16 and during 3D training it was reduced to 1 due to memory constraints. In order to compensate for the ineffectiveness of the batch normalization modules with batch size 1, we used SeLu activations, recently introduced in [7]. These were found to very effectively accelerate convergence. The models were optimized using standard stochastic gradient descent with learning rate fixed at 0.001 and momentum set to 0.9. The data set was split in 80% training, 10% validation, 10% testing. The models were trained in turn on an NVidia DGX-1 with 8X P100 GPUs (16 Gb RAM per GPU), with whole model \(8\times \) replication (except for the CNN, which was trained on 4 GPUs). The models were pre-trained for 20 epochs and trained for 50 epochs. Training time was 4.6 h for the UNet and 86 h for the YNet. Quantitative results are reported in Table 1. Examples of image segmentations produced by the UNet and the YNet are reported in Fig. 3. The YNet determines an increase of Dice score from 0.581 to 0.623 on the test set, and an improvement in quality of the segmentations. The improvement is visible in particular in small lesions and in the reduction of artifacts. The UNet outperformed the CNN in classification, achieving sensitivity 0.981 and specificity 0.980.
5 Discussion and Conclusion
In this paper, we have introduced a deep learning architecture to automatically detect and segment acute ischemic stroke lesions on diffusion-weighted MRI to address the challenges of stroke diagnosis and optimal selection of patients for acute stroke therapy. The model is validated on a very large clinical dataset and achieves state-of-the-art performance. To the best of our knowledge, our approach to classify and segment a medical image from both pixel level annotations and weak image level labels and using 3D contextual information is novel. By using end-to-end multi-task learning, the model achieves high classification and segmentation performance, learning to segment from weak labels and to classify from manual annotations. Given that many problems in image interpretation require detection and localization, we believe that the approach explored in this paper can have wide applicability in medical image interpretation, enabling machine learning at large scale with minimum annotation effort. In the future, this method could be extended to account for multiple segmentation and classification labels.
References
Berkhemer, O., et al.: A randomized trial of intraarterial treatment for acute ischemic stroke. N. Engl. J. Med. 372(1), 11–20 (2015)
Chen, L., Bentley, P., Rueckert, D.: Fully automated acute ischemic lesion segmentation in DWI using convolutional neural networks. Neuroimage 15, 633–643 (2017)
Çiçek, Ö., Abdulkadir, A., Lienkamp, S.S., Brox, T., Ronneberger, O.: 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 424–432. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_49
Glorot, X., Bengio, Y.: Understanding the difficulty of training deep feedforward neural networks. In: PMLR, vol. 9, pp. 249–256 (2010)
Goyal, M., Menon, B., van Zwam, W.: Endovascular thrombectomy after large-vessel ischaemic stroke: a meta-analysis of individual patient data from five randomised trials. Lancet 387, 1723–1731 (2016)
Hacke, W.: A new DAWN for imaging-based selection in the treatment of acute stroke. N. Engl. J. Med. 378, 81–83 (2018)
Klambauer, G., Unterthiner, T., Mayr, A.: Self-normalizing neural networks. In: NIPS, pp. 971–980 (2017)
Martel, A.L., Allder, S.J., Delay, G.S., Morgan, P.S., Moody, A.R.: Measurement of infarct volume in stroke patients using adaptive segmentation of diffusion weighted MR images. In: Taylor, C., Colchester, A. (eds.) MICCAI 1999. LNCS, vol. 1679, pp. 22–31. Springer, Heidelberg (1999). https://doi.org/10.1007/10704282_3
Milletari, F., Navab, N., Ahmadi, S.: V-Net: fully convolutional neural networks for volumetric medical image segmentation. In: 3DV, pp. 565–571 (2016)
Papandreou, G., Chen, L., Murphy, K., Yuille, A.: Weakly- and semi-supervised learning of a DCNN for semantic image segmentation. CoRR 1502.02734 (2015)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Subudhi, A., Jena, S., Sabut, S.: Delineation of the ischemic stroke lesion based on watershed and relative fuzzy connectedness in brain MRI. Med. Biol. Eng. Comput. 56, 1–13 (2017)
Tsai, J., et al.: Automated detection and quantification of acute cerebral infarct by fuzzy clustering and histographic characterization on diffusion weighted MR imaging and apparent diffusion coefficient. BioMed. Res. Int. (2014). Article no. 963032
Tustison, N.: N4ITK: improved N3 bias correction. IEEE Trans. Med. Imaging 29(6), 1310–1320 (2010)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Pedemonte, S. et al. (2018). Detection and Delineation of Acute Cerebral Infarct on DWI Using Weakly Supervised Machine Learning. In: Frangi, A., Schnabel, J., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science(), vol 11072. Springer, Cham. https://doi.org/10.1007/978-3-030-00931-1_10
Download citation
DOI: https://doi.org/10.1007/978-3-030-00931-1_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00930-4
Online ISBN: 978-3-030-00931-1
eBook Packages: Computer ScienceComputer Science (R0)