Abstract
Recent CNN architectures have established state-of-the-art results in a large range of medical imaging applications. We propose an extension to the U-Net architecture relying on multi-task learning: while keeping a single encoding module, multiple decoding modules are used for concurrent segmentation tasks. We propose improvements of the encoding module based on the latest CNN developments: residual connections at every scale, mixed pooling for spatial compression and large kernels for convolutions at the lowest scale. We also use dense connections within the different scales based on multi-size pooling regions. We use this new architecture to jointly detect and segment red and bright retinal lesions which are essential biomarkers of diabetic retinopathy. Each of the two categories is handled by a specialized decoding module. Segmentation outputs are refined with conditional random fields (CRF) as RNN and the network is trained end-to-end with an effective Kappa-based function loss. Preliminary results on a public dataset in the segmentation task on red (resp. bright) lesions shows a sensitivity of 66,9% (resp. 75,3%) and a specificity of 99,8% (resp. 99,9%).
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
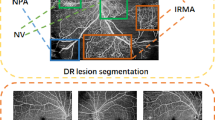
Diabetic retinopathy (DR) is a potential consequence of diabetes, affecting nearly 34% of the diabetic population. The disease progresses through stages characterized mainly by the lesions observed in the retina. In 2D fundus, those lesions can be regrouped in two categories according to their appearance: bright (such as exudates and cotton wool spots) and red (such as hemorrhages and microaneurysms). Most of the literature on DR lesion segmentation proposes three main stages: candidates detection, candidates classification and refinement of the segmentation. This approach is used for example for red lesion detection in [1] using handcrafted features. Deep learning has been used in [2] for hemorrhage detection. For bright lesions, detection and segmentation usually rely on unsupervised methods, as in [3]. Nonetheless, clinical assessment requires detection of all types of lesions. This hypothesis stems from the empirical observation of the labeling process done by medical experts on fundus images. Each salient region is classified according to its specific content but also to the context of the entire image (like the presence of other lesions). An automatic decision system can learn implicitly the DR grading by using a model as a black box. However, to reproduce the protocol used by the grader the decision should rely on an explicit full detection of lesions. Meanwhile, the capacity of CNNs to segment medical images obtained from multiple modalities through multitasking has been demonstrated in [4]. Even for a single modality, multitasking is well suited for jointly segmenting different types of lesions. This approach provides several advantages, especially shorter inference times and the ability to train a single architecture to independently perform multiple highly specialized tasks that share a common basis.
To our knowledge, there are no methods based on fully convolutional approaches used for joint lesions segmentations. To address this gap, this paper focuses on segmenting bright and red lesions with a single deep multitask architecture, without the need of blood vessels nor optic disc removal. We propose a novel network based on recent developments of CNNs, like Residual Connections, Global Convolution and Mixed-pooling. We also introduce Dense Pooling Connections, a new type of connection that is designed to reinforce the robustness to noise by aggregating maximum activations within multiple regions. We prove a performance improvement in comparison with existing architecture.
2 Methods
Overview. We train a novel CNN architecture with patches randomly extracted from normalized images. The architecture extends the U-Net [5] with multi-task learning. Improvement of the descending part (the encoder) of U-Net is proposed as well as a new training strategy. The features from the encoder are shared by two decoders respectively specialized respectively in bright and red lesions segmentation.
The network is fed patches from the normalized images. (a) The encoding module uses a generic set of parameters shared by the two tasks. (b) The decoding modules are task-specific. An auxiliary cost (c) is added at the end of the encoding module; it is trained only to predict the presence of lesions.
2.1 Multitask Architecture
Multitask learning was introduced in [6] as a way to improve generalization. Part of the model is shared across independent tasks, while each task has its specifics parameters. Figure 1 shows our global architecture and Fig. 2 describes the encoding module in detail. The intuition behind multi-task learning in our case is that information needed for bright and red lesions segmentation is common to both tasks (for example, anatomical features of the retina).
Mixed Pooling: Each max-pooling is replaced by mixed-pooling layer [7]. For an input tensor \(\varvec{x}\) composed of N channels and a vector \(\varvec{a}\) (trainable parameter), the mixed-pooling layer computes:
We use one scalar (\(\varvec{a}_n \in [0,1]\)) per layer/channel, for an efficient combination without drastically increasing the number of parameters of the model (N additional parameters per pooling layer).
Residual Connection: At each resolution level, the two \(3\times 3\) convolutions of the original U-Net are extended to become residual blocks as introduced in [8]. The motivation is to prevent the degradation problem observed in large models by allowing the blocks to possibly become identity mappings.
Dense Pooling Connections: We introduce dense pooling connections through multiple resolution levels. Each level is connected to those beneath it. Pooling operations with various pooling sizes guarantee spatial resolution consistency. We make the hypothesis that pooling operations over successively larger regions reinforce scale and translation invariance while reducing sensitivity to noise as more and more context is added. At the lowest level, for a given field of view, every previous levels transmit a combination of its maximal and average activation. The aggregation of those data should facilitate discrimination between relevant features and local noise.
Global Convolutional Network: At the lowest scale of the network, we use convolutions with large kernels following the implementation recently proposed in [9]. This further aggregates the contextual information.
Task Specific Decoders. The decoding modules used are the same as in the original U-Net design. We use two decoding modules, each specialized for one lesion category. Near the end of the training, we also added two fully connected Conditional Random Fields (CRFs). CRFs were originally introduced by [10]. We use the softmax output of each decoding module as the unary potential. The pair-wise potential consists in a weighted sum of two Gaussian kernels that “control” the appearance and the smoothness of the segmentation. The parameters of the kernels are trained with the rest of the network, according to the proposed method in [11] which implements the CRF as an additional RNN layer on top of a traditional convolutional architecture.
2.2 Training
Each task is associated with its specific cost function. We also use an auxiliary cost trained to detect whether a lesion is present or not in the patch. This helps the encoding module to focus on distinguishing between an actual lesion and other biomarkers. During training, the objective function \(C_{global}\) is the weighted sum of each cost:
Training is performed in three stages. In the first stage, the network is trained with a log-likelihood based cost (\(\mathcal {L}(\theta \mid x) = -\frac{1}{D}\sum _i^{D} \log P(Y=y^{(i)}|x^{(i)}, \theta )\)).
In the second stage, we change the objective function to a Kappa-based one. Cohen’s Kappa (\(\kappa \)) coefficient measures the agreement between two raters. As it takes into account the possibility of agreement occurring by chance, this coefficient is well suited for distinguishing highly unbalanced classes as in our case. The core idea of the \(\kappa \) coefficient is to quantify the difference between the accuracy \(\rho _{acc}\) and the probability of pure chance agreement \(\rho _{chance}\):
As the accuracy is not a differentiable measure, we use soft approximation to model it. The output of the softmax, \(y_{proba}\), approximates the predicted label, \(y_{pred}\), which is valid for high-confidence predictions as \(y_{proba}\) tends to \(y_{pred}\) encoded in a one-hot vector. This is why we initially train the network with the likelihood \(\mathcal {L}\), in order to obtain this high level of confidence.
The third training stage adds the CRFs after the two decoders. The auxiliary cost is discarded and only the weights of the two decoding modules are updated. Table 1 summarizes the parameters for the training stages.
As an optimizer, we use the Adadelta algorithm introduced in [12]. The weights update policy is:
Where \(E[\varDelta x^2]\) and \(E[g^2]\) are the running averages characterized by a parameter \(\gamma \). We use \(\gamma =0.95\) (a high value counter-balances the noise introduced by small batch sizes). As Adadelta is designed to remove the need of an explicit learning rate, in the original paper [12] \(\nu \) is fixed and equal to 1. Nonetheless, as it was also originally suggested, we found that dividing \(\nu \) by 10 every 20 epochs drastically helps the convergence, as shown in Fig. 3.
3 Experiments
We mainly used the publicly available DIARETDB1 database [13], which provides 89 fundus images from DR patients. As this database was designed for lesion detection rather than segmentation, we refined the lesions boundaries manually, and an ophthalmologist validated them. 61 images were used for test and validation (8 images from the recommended test set were randomly selected for the validation set). The training set was composed of 28 images from DIARETDB1, supplemented by 17 images with lesions from a private database and 18 healthy images extracted from the e-ophtha database [14], giving a total of 63 training images. A simple preprocessing step was applied to normalize the illumination and we increased the dataset using data augmentation. We applied geometrical (translation, rotation, shearing and elastic distortion) and color (brightness, contrast, gamma, HSV saturation/value) transformations to the input images. For each image, a random combination of those operations was applied. The parameters of each transformation were also randomly sampled at each epoch. We thereby ensured that the network never saw the exact same patch twice. The network was fed patches of size \(400\times 400\). Between 8 and 10 patches were randomly extracted per image, with a prior distribution to favor patches centered on a lesion. We used a weights decay rate of 0.0005 and a batch size of 2.
4 Results and Discussion
We tested our model by comparing it with the original U-Net architecture (one decoder, three classes), and with another model similar to the U-Net but with two decoding modules. We refer to these latter networks as U-Net and U-Net2; we trained them with the same strategy as our proposed network. Sensitivity and specificity were measured pixel-wise and averaged over the test set. Tables 2 and 3 provide the segmentation performance results. The quality of the segmentation was also evaluated in a patch-wise manner, as this corresponds to what the network actually “sees”. Patches were of size \(400\times 400\). We averaged the \(\kappa \) and the Dice coefficient s, measured per patch, to get averages per image \(\kappa _{image}\) and \(s_{image}\). To get global values, we then averaged each \(\kappa _{image}\) and \(s_{image}\) over the entire test set (see Tables 4 and 5).
The results are encouraging with regard to the proposed network’s segmentation performance in comparison with both U-Net and U-Net2. The U-Net gave satisfactory results in bright lesions segmentation but was completely unable to predict red lesions. This gives strong support in favor of multitasking, as specialized branches appear to be able to capture features that a single branch cannot (at least for the same number of training epochs). Nonetheless, we also observe that our results tend to globally worsen with the CRFs. Visual inspection shows that the CRFs tend to add tiny false positive red lesions, near the vessels. In addition, the CRFs are well suited for hard exudates but tend to miss the boundaries of soft ones. The inference time was approximately 1 s per image, running on NVIDIA GTX 1070 Ti hardware. Obtaining a fast and complete segmentation of the image constitutes an important first step toward our ultimate goal of constituting an extensive, fully labelled fundus image database. This process will be greatly accelerated using our model. We also plan to assess the capacity of grading DR using features obtained directly from the encoding module and output segmentation results. Indeed, the inferior results of the basic U-Net as compared to the multi-task networks suggests that in those, encoded features are highly representative of the abnormalities observed in the images.
5 Conclusion
We have proposed a novel CNN architecture to jointly segment bright and red lesions in fundus images. We have highlighted the value of a multitask learning approach, as opposed to single task classification. The present work opens the door to many possibilities, from clinical assistance (computer-assisted lesion identification) to DR grading methods that do not rely on a “black-box” approach.
References
Seoud, L., Hurtut, T., Chelbi, J., Cheriet, F., Langlois, J.M.P.: Red lesion detection using dynamic shape features for diabetic retinopathy screening. IEEE Trans. Med. Imaging 35(4), 1116–1126 (2016)
van Grinsven, M.J.J.P., van Ginneken, B., Hoyng, C.B., Theelen, T., Sánchez, C.I.: Fast convolutional neural network training using selective data sampling: application to hemorrhage detection in color fundus images. IEEE Trans. Med. Imaging 35(5), 1273–1284 (2016)
Vanithamani, R., Renee Christina, R.: Exudates in detection and classification of diabetic retinopathy. In: Abraham, A., Cherukuri, A.K., Madureira, A.M., Muda, A.K. (eds.) SoCPaR 2016. AISC, vol. 614, pp. 252–261. Springer, Cham (2018). https://doi.org/10.1007/978-3-319-60618-7_25
Moeskops, P., et al.: Deep learning for multi-task medical image segmentation in multiple modalities. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 478–486. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_55
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Caruana, R.: Multitask learning. Mach. Learn. 28(1), 41–75 (1997)
Yu, D., Wang, H., Chen, P., Wei, Z.: Mixed pooling for convolutional neural networks. In: Miao, D., Pedrycz, W., Ślȩzak, D., Peters, G., Hu, Q., Wang, R. (eds.) RSKT 2014. LNCS (LNAI), vol. 8818, pp. 364–375. Springer, Cham (2014). https://doi.org/10.1007/978-3-319-11740-9_34
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 770–778, June 2016
Peng, C., Zhang, X., Yu, G., Luo, G., Sun, J.: Large kernel matters - improve semantic segmentation by global convolutional network. CoRR abs/1703.02719 (2017)
Krähenbühl, P., Koltun, V.: Efficient inference in fully connected CRFs with Gaussian edge potentials. CoRR abs/1210.5644 (2012)
Zheng, S., et al.: Conditional random fields as recurrent neural networks. In: 2015 IEEE International Conference on Computer Vision (ICCV), pp. 1529–1537, December 2015
Zeiler, M.D.: ADADELTA: an adaptive learning rate method. CoRR abs/1212.5701 (2012)
Kauppi, T., et al.: DIARETDB1 diabetic retinopathy database and evaluation protocol (01 2007)
Decenciàre, E., et al.: TeleOphta: machine learning and image processing methods for teleophthalmology. IRBM 34(2), 196–203 (2013). Special issue: ANR TECSAN: Technologies for Health and Autonomy
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Playout, C., Duval, R., Cheriet, F. (2018). A Multitask Learning Architecture for Simultaneous Segmentation of Bright and Red Lesions in Fundus Images. In: Frangi, A., Schnabel, J., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science(), vol 11071. Springer, Cham. https://doi.org/10.1007/978-3-030-00934-2_12
Download citation
DOI: https://doi.org/10.1007/978-3-030-00934-2_12
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00933-5
Online ISBN: 978-3-030-00934-2
eBook Packages: Computer ScienceComputer Science (R0)