Abstract
Most whole-slide histological images are stained with hematoxylin and eosin dyes. Slide stain separation or color deconvolution is a crucial step within the digital pathology workflow. In this paper, the blind color deconvolution problem is formulated within the Bayesian framework. Our model takes into account both spatial relations among image pixels and similarity to a given reference color-vector matrix. Using Variational Bayes inference, an efficient new blind color deconvolution method is proposed which provides a fully automated procedure to estimate all the unknowns in the problem. A comparison with classical and current state-of-the-art color deconvolution algorithms, using real images with known ground truth hematoxylin and eosin values, has been carried out demonstrating the superiority of the proposed approach.
This work was supported in part by the Spanish Ministerio de Economía y Competitividad under contract DPI2016-77869-C2-2-R.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
In digital brightfield microscopy, tissues are usually stained before digitization and evaluation, with hematoxylin and eosin (H&E) being the most widely used stains. Color deconvolution (CD) aims at separating a color image into the concentration of each stain present in it. This is not an easy task since the exact spectral profile of the stains varies from one image to another [10]. Hence, the stain color-vector matrix, which relates the color image and the stain concentrations, often needs to be estimated for each slide. Once the stain color-vectors are calculated, the color of different images can be normalized to a target image for an easier evaluation. This is usually done by replacing the stain vectors with the target stain vectors obtained from the reference slide, and converting the calculated concentrations back to an RGB image.
One of the first CD methods was proposed by Ruifrok et al. [9]. The values for the stain vector of each dye were obtained by measuring the relative absorption of each color from single-stained images. While these standard values are frequently used, they have a strong dependence on the experimental setting utilized in [9]. In [6] H&E color-vector are obtained from the two largest singular values of the SVD decomposition of the optical density matrix. McCann et al. [7] extend the method in [6] by adjusting the contrast of the eosin channel and including a weak interaction between eosin and hematoxylin in the pixels of the hematoxylin channel where eosin values were changed. The algorithm is tested on a set of three H&E images that were stained and destained to create H-only and E-only images that are used as ground-truth separated images for the H&E image. In [4] the stain color-vectors are estimated by projecting the input color image in the Maxwellian chromaticity plane to form clusters, each one corresponding to one stained tissue type. In [5] color normalization is performed by first deconvolving both source and target images, applying a non-linear mapping of the source to the target image channels and recombining the mapped channels into the normalized source image. In [8] the CD problem is formulated as a blind source separation problem tackled by Non-negative Matrix Factorization (NMF) and Independent Component Analysis (ICA). Vahadane et al. [11] and Xu et al. [13] independently extend the NMF in [8] with regularization and sparsity terms since they assume that most pixels contain one type of biological material. The use of Non-negative Least Squares (NNLS) instead of NMF is proposed in [3]. Alsubaie et al. [1] propose using ICA in the wavelets domain where the independence condition among sources is relaxed. All these methods depend on parameters that need to be manually adjusted for an optimal deconvolution.
In this paper we propose a novel fully automated CD method that simultaneously estimates the color-vector matrix, the concentration of the stains, and all required model parameters. In our Bayesian blind CD problem formulation we introduce a smoothness prior model on the stain concentrations which helps reduce the acquisition noise and takes into account the spatial correlation between adjacent pixels. Despite the variability among images, the color-vector matrices are often assumed to be close to a commonly accepted standard matrix. Our Bayesian modelling allows us to include this additional prior knowledge on the sought after solution.
The rest of the paper is organized as follows: in Sect. 2 we mathematically formulate the blind CD of histopathological images problem. This problem is approached using the Bayesian framework in Sect. 3. In this section we also carry out Bayesian inference to estimate the color-vector matrix, concentrations, and model parameters in a fully automated manner. In Sect. 4 the proposed method is evaluated in a set of H&E stained images and its performance is compared with other classical and state-of-the-art CD methods. Finally, Sect. 5 concludes the paper.
2 Problem Formulation
The RGB intensity image detected by a brightfield microscope observing a stained histological specimen’s slide is the \((M\times N)\times 3\) matrix, \({\mathbf {I}}\), with columns \({\mathbf {i}}_c=(i_{1c},\ldots ,i_{MNc})^\mathrm{T}, c\in \{R,G,B\}\) and MN the number of pixels. According to the monochromatic Beer-Lambert law [9], the Optical Density (OD) for channel c of the slide, \({\mathbf {y}}_c \in \mathbb {R}^{MN\times 1}\), is \({\mathbf {y}}_c = - \log _{10}\left( {{\mathbf {i}}_c}/{{\mathbf {i}}^{0}_c}\right) \), where \({\mathbf {i}}^{0}_c\) denotes the incident light, and the division operation and \(\log _{10}(\cdot )\) function are computed element-wise. For a slide stained using \(n_s\) stains the observed OD image \({\mathbf {Y}}={[{\mathbf {y}}_R,{\mathbf {y}}_G,{\mathbf {y}}_B]} \in \mathbb {R}^{MN\times 3}\) can be obtained from
where \({\mathbf {N}}\) is the capture noise matrix of size \(MN\times 3\) with i.i.d. \(\mathcal{N}(0,\beta ^{-1})\) components, \({\mathbf {C}}\in \mathbb {R}^{MN\times n_s}\) is the stain concentration matrix
with i-th row \({\mathbf {c}}_{i,:}^\mathrm{T}=(c_{i1},\ldots ,c_{in_s})\), \(i=1,\ldots ,MN\) and columns \({\mathbf {c}}_s=(c_{1s},\ldots ,\) \(c_{MNs})^\mathrm{T}\), \(s\in \{1,\ldots ,n_s\}\) and \({\mathbf {M}}\in \mathbb {R}^{3\times n_s}\) is the normalized stains’ specific color-vector matrix. Each column in matrix \({\mathbf {M}}\) is a unit \(\ell 2\) norm stain color-vector containing the relative RGB color composition of the corresponding stain. Color Deconvolution (CD) is a technique that allows to obtain the stain concentration matrix, \({\mathbf {C}}\), and the color-vector matrix, \({\mathbf {M}}\), from the observed optical densities, \({\mathbf {Y}}\). In the following section we will use Bayesian modeling and inference to estimate \({\mathbf {M}}\) and \({\mathbf {C}}\) as well as the model parameters.
3 Bayesian Modelling and Inference
Following the degradation model in (1), we have
The stain concentrations at each pixel on the image are expected to have values similar to the ones of the surrounding pixels. So, we impose smoothing prior models on the concentrations \({\mathbf {c}}_s\), \(s=1,\ldots ,n_s\), that is, on the columns of \({\mathbf {C}}\), as the Gaussian distributions of the form
where \({\mathbf {F}}\in \mathbb {R}^{MN\times MN}\) is a smoothing filter and \(\alpha _s\), \(s=1,\ldots ,n_s\), controls the amount of smoothness.
The color-vector matrix \({\mathbf {M}}=[{\mathbf {m}}_1,\ldots ,{{\mathbf {m}}_{n_s}}]\) is also unknown, because it depends on the staining procedures and microscopes. In [9], standard color-vectors for hematoxylin, eosin, and DAB stains were proposed. Although those standard color-vectors are not usually exact for each single image, they are very representative and have been frequently used. In this paper we incorporate the similarity to a representative color-vector matrix \(\underline{{\mathbf {M}}}=[\underline{{\mathbf {m}}}_1,\ldots ,{\underline{{\mathbf {m}}}_{n_s}}]\) into the Gaussian prior model
where \(\gamma _s\), \(s=1,\ldots ,n_s\), controls our confidence on the accuracy of \(\underline{{\mathbf {m}}_s}\).
The joint probability distribution for our problem is
Following the Bayesian paradigm, inference will be based on the posterior distribution \({\mathrm p}({\mathbf {C}},{\mathbf {M}},\beta ,{\varvec{\alpha }},{\varvec{\gamma }}|{\mathbf {y}})\) which cannot be obtained in closed form, so a variational approach [2] is applied.
The hyperparameters \(\{\beta ,{\varvec{\alpha }},{\varvec{\gamma }}\}\) have not been considered as variables for the variational method, but as model parameters for which a Maximum Likelihood Estimator (MLE) will be obtained and \({\mathrm p}({\mathbf {C}},{\mathbf {M}},\beta ,{\varvec{\alpha }},{\varvec{\gamma }}|{\mathbf {y}})\) is approximated by the distribution
It can then be shown [2] that for each unknown \(\theta \in \varTheta =\{{\mathbf {m}}_1,\ldots ,{\mathbf {m}}_{n_s}\), \({\mathbf {c}}_1,\ldots ,{\mathbf {c}}_{n_s}\}\), \({\mathrm q}(\theta )\) will have the form
where \(\varTheta {\setminus }\theta \) represents all the variables in \(\varTheta \) except \(\theta \) and \(\left\langle \cdot \right\rangle _{{\mathrm q}(\varTheta {\setminus }\theta )}\) denotes expected value calculated using the distribution \({\mathrm q}(\varTheta {\setminus }\theta )\). Estimates for the different variables can be obtained as \(\hat{\theta }= {\left\langle \theta \right\rangle }_{{\mathrm q}(\theta )}\). Let us now obtain the analytic expressions for each unknown estimate.
Concentration Update. Let us define
which produces \({\mathrm q}({\mathbf {c}}_s) = {\mathcal {N}}({\mathbf {c}}_s|\left\langle {\mathbf {c}}_s\right\rangle ,{\varvec{\varSigma }}_{{\mathbf {c}}_s})\,,\) where
Color-Vector Update. In a similar way, using Eq. (9), from Eqs. (6) and (8) we now have
which produces \({\mathrm q}({\mathbf {m}}_s) = {\mathcal {N}}({\mathbf {m}}_s|\left\langle {\mathbf {m}}_s\right\rangle ,{\varvec{\varSigma }}_{{\mathbf {m}}_s})\,,\) where
Notice that \(\left\langle {\mathbf {m}}_s\right\rangle \) may not be an unitary vector. We can always replace \(\left\langle {\mathbf {m}}_s\right\rangle \) by \(\left\langle {\mathbf {m}}_s\right\rangle /\parallel \left\langle {\mathbf {m}}_s\right\rangle \parallel \) and \({\varvec{\varSigma }}_{{\mathbf {m}}_s}\) by \({\varvec{\varSigma }}_{{\mathbf {m}}_s}/\parallel \left\langle {\mathbf {m}}_s\right\rangle \parallel ^2\). Notice also that \(\left\langle c_{is}^2\right\rangle \) can be calculated using (11) and \(\left\langle \parallel {\mathbf {m}}_s\parallel ^2\right\rangle \) can be easily calculated from (13).
Parameter Update. Finally, the MLE estimators of the noise, concentration, and color-vectors precisions are
respectively. Notice that the involved expected values can be easily calculated.

The proposed Variational Bayesian Blind Color Deconvolution method, summarized in Algorithm 1, allows to obtain the estimated concentrations \(\hat{{\mathbf {c}}}_s\) and color-vector \(\hat{{\mathbf {m}}}_s\) iterating on the concentration and color-vector updates until convergence. Finally, an RGB image of each separated stain, \(\hat{{\mathbf {i}}}^{\mathrm {sep}}_s\), can be obtained as \(\hat{\mathbf {i}}^{\mathrm {sep}}_s = \exp _{10}({-\hat{{\mathbf {m}}}_s \hat{{\mathbf {c}}}_s^{\mathrm {T}}})\).
4 Experiments
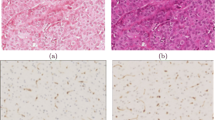
We compared the proposed fully automated approach with classical and state-of-the-art CD methods on the stain separation benchmark in [7]. This dataset is formed by three H&E images and their corresponding H-only and E-only images that can be used as ground truth images for the color deconvolution procedure. Each image in the dataset was obtained by eosin staining the tissue, imaging, destaining, staining with hematoxylin, imaging, staining also with eosin and imaging. An example of H&E stained image in the dataset is shown in Fig. 1(a) and its corresponding E-only and H-only images are shown in the left and right hand side of Fig. 1(b), respectively.
The election of a reference color-vector matrix depends on the used stains. For the H&E stains used in this paper, the value of \(\underline{{\mathbf {M}}}\) was set to the H&E values proposed in [9]. We want to note that, for stains different from H&E, simply measuring the tissue response to a single stain might provide its corresponding reference color-vector. The convergence criterion \(\Vert \left\langle {\mathbf {c}}_s\right\rangle ^{(n)}- \left\langle {\mathbf {c}}_s\right\rangle ^{(n-1)}\Vert ^2/\Vert \left\langle {\mathbf {c}}_s\right\rangle ^{(n)}\Vert ^2 < 10^{-5}\) for both stains, that is, \(s=1,2\), was used. This is met in about 15 iterations of the algorithm. We compare against the classical method in [6] and the recent methods in [7, 11]. For all the competing algorithms, parameters were selected following the recommendations on the original paper or the reference software freely available.
Figure 1(c)–(f) shows the separations with the proposed and competing methods. From the images, it is clear that the method in [7] and the proposed algorithm produce results closer to the ground truth (see Fig. 1(b)) than the methods in [6, 11]. Although all methods effectively separate epithelial and stromal structures, ours seems closer to ground truth. Note, for instance, that the long structure in the center of the image (corresponding to bone tissue [7]) is not clearly shown in the hematoxylin estimations in Figs. 1(c) and (e). All eosin estimations present a higher contrast than the ground truth although, estimations obtained by the proposed method and the method in [7] are more similar to the ground truth. The eosin estimation from the method in [7] seems to be slightly less contrasted than ours.
Numerical results, using the Peak Signal to Noise Ratio (PSNR) and Structural Similarity (SSIM) [12] measures, are presented in Table 1. This table includes the results for the non-blind method in [9] as a reference. The figures-of-merit confirm the visual inspection results. The proposed method performs better than the competitors, except for the case of the eosin stain for the algorithm in [7]. This was expected since this algorithm selectively modifies the obtained values for the stain separations to better accommodate ground truth. More precisely, in [7] the eosin separation is corrected in contrast by adding a small part of the hematoxylin stain, and the hematoxylin stain is then computed again by taking into account interaction between the stains in those places where the contrast of the eosin coefficients is adjusted. Note that, in spite of these adjustments, our fully automated proposed method consistently provides better PSNR results for the hematoxylin stain than the method in [7]. Notice that the obtained PSNR and, especially, the SSIM values are quite low. This is due to the staining-destaining process that makes the tissue to move and deform. These deformations were partially corrected in [7] by geometrically registering the H-only and E-only to their corresponding estimations. Although we used the registered images from [7] as ground truth for all the tests, there still are misalignment between ground truth and estimations that deteriorate the figures-of-merit.
The proposed method is faster than the recent method in [11], taking 14 s on a i7-5550U @ 2.40 GHz laptop with 16 GB RAM versus 50.2 s. However it is slower than the classical method in [6], that took 0.4 s, and the method in [7] that took 2.78 s.
5 Conclusions
A novel fully automated variational Bayesian blind color deconvolution method for histological images is proposed. The method estimates the color-vector matrix, the concentration of the stains and all the model parameters. The proposed model takes into account the spatial relations between pixels as well as the similarity to a standard color-vector matrix. Comparison with classical and recent methods demonstrated that the proposed method produces better results than the competitors, except for the eosin stain by the algorithm in [7] as already mentioned.
References
Alsubaie, N., Trahearn, N., et al.: Stain deconvolution using statistical analysis of multi-resolution stain colour representation. PLOS ONE 12(1), e0169875 (2017)
Bishop, C.: Pattern Recognition and Machine Learning, pp. 454–455. Springer, New York (2006)
Carey, D., Wijayathunga, V.N., Bulpitt, A.J., Treanor, D.: A novel approach for the colour deconvolution of multiple histological stains. In: Proceedings of the 19th Conference of Medical Image Understanding and Analysis, pp. 156–162 (2015)
Gavrilovic, M., Azar, J.C., et al.: Blind color decomposition of histological images. IEEE Trans. Med. Imaging 32(6), 983–994 (2013)
Khan, A.M., Rajpoot, N., Treanor, D., Magee, D.: A nonlinear mapping approach to stain normalization in digital histopathology images using image-specific color deconvolution. IEEE Trans. Biomed. Eng. 61(6), 1729–1738 (2014)
Macenko, M., Niethammer, M., et al.: A method for normalizing histology slides for quantitative analysis. In: 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, pp. 1107–1110 (2009)
McCann, M.T., Majumdar, J., et al.: Algorithm and benchmark dataset for stain separation in histology images. In: IEEE International Conference on Image Processing (ICIP), pp. 3953–3957 (2014)
Rabinovich, A., Agarwal, S., Laris, C., Price, J.H., Belongie, S.J.: Unsupervised color decomposition of histologically stained tissue samples. In: Advances in Neural Information Processing Systems, pp. 667–674 (2004)
Ruifrok, A.C., Johnston, D.A.: Quantification of histochemical staining by color deconvolution. Anal. Quant. Cytol. Hist. 23(4), 291–299 (2001)
Taylor, C.R., Levenson, R.M.: Quantification of immunohistochemistry-issues concerning methods, utility and semiquantitative assessment II. Histopathology 49(4), 411–424 (2006)
Vahadane, A., Peng, T., et al.: Structure-preserving color normalization and sparse stain separation for histological images. IEEE Trans. Med. Imaging 35(8), 1962–1971 (2016)
Wang, Z., Bovik, A.C., et al.: Image quality assessment: from error visibility to structural similarity. IEEE Trans. Image Proc. 13, 600–612 (2004)
Xu, J., Xiang, L., Wang, G., et al.: Sparse Non-negative Matrix Factorization (SNMF) based color unmixing for breast histopathological image analysis. Comput. Med. Imaging Graph. 46, 20–29 (2015)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Hidalgo-Gavira, N., Mateos, J., Vega, M., Molina, R., Katsaggelos, A.K. (2018). Fully Automated Blind Color Deconvolution of Histopathological Images. In: Frangi, A., Schnabel, J., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science(), vol 11071. Springer, Cham. https://doi.org/10.1007/978-3-030-00934-2_21
Download citation
DOI: https://doi.org/10.1007/978-3-030-00934-2_21
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00933-5
Online ISBN: 978-3-030-00934-2
eBook Packages: Computer ScienceComputer Science (R0)