Abstract
This paper proposes a weakly-supervised representation learning framework for probe-based confocal laser endomicroscopy (pCLE). Unlike previous frame-based and mosaic-based methods, the proposed framework adopts deep convolutional neural networks and integrates frame-based feature learning, global diagnosis prediction and local tumor detection into a unified end-to-end model. The latent objects in pCLE mosaics are inferred via semantic label propagation and the deep convolutional neural networks are trained with a composite loss function. Experiments on 700 pCLE samples demonstrate that the proposed method trained with only global supervisions is able to achieve higher accuracy on global and local diagnosis prediction.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Probe-based confocal laser endomicroscopy (pCLE) is a novel optical biopsy technique for real-time tissue characterization in vivo. Flexible coherent fiber-bundle probes, typically of the size of 1.0 mm in outer diameter, integrating confocal optics in the proximal end, are used to provide fluorescence imaging of the biological tissue. Furthermore, these miniaturized probes can be integrated into standard video endoscopes, making pCLE a popular choice for minimally invasive endoscopic procedures. Current applications include breast, gastro-intestinal and lung diseases.
Although pCLE enables the acquisition of in-vivo microscopic images that resemble the gold-standard (H&E) stained histology images, many challenges associated with disease characterization still remain. A major challenge being that the field of view (FOV), limited by the size of the fiber bundle, is typically less than 1 mm\(^2\). A high resolution Cellvizio probe for example offers a lateral resolution of 1.4 \(\upmu \)m but a FOV of just 240 \(\upmu \)m. With such a small FOV, particularly when compared to histology slides, means that only a small number of morphological features can be visualized in each image. Furthermore, conventionally histology images are examined by trained pathologists, which is different from the surgical setting where live pCLE images need to be assessed in real-time by surgeons who may have limited training on histopathology.
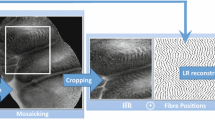
For these reasons, there has been extensive interests in developing computer-aided diagnosis for automated pCLE image classification in the recent years [1,2,3,4,5]. These methods can be broadly categorized into frame-based methods [2, 3, 5] and mosaic-based methods [1, 4]. As shown in Fig. 1(a), the frame-based methods adopt the visual information of single frame based on Dense-SIFT [2], deep convolutional neural networks (CNN) [5] or transfer learning from histopathological images [3]. Although these methods provide diagnosis result for each pCLE frame, the FoV of each frame is relatively small leading to low confidence for final diagnosis. Moreover, the frame-based methods require massive annotations of training data which is often expensive and time-consuming. On the other hand, the mosaic-based methods [1, 4], as shown in Fig. 1(b), help to increase the effective FoV along with direction of probe motion by stitching consecutive image frames. Even with this enlarged FoV, the pCLE diagnostic accuracy depends on the quality of reconstructed mosaics, which in turn is affected by several factors including the speed by which the operator can translate the probe across the tissue, as well as probe-tissue contact and tissue deformation. In addition, mosaic-based methods can only provide a global diagnosis for the large pCLE mosaic but not for the specific regions of the mosaic (e.g. for the regions that correspond to neoplastic tissue as shown in Fig. 1(c)). This would affect the overall diagnostic performance. To provide an objective support for pCLE diagnosis, it is critical to develop a learning framework that can provide both global diagnosis as well as local tumour detection for pCLE images.
Given only the global diagnosis of the pCLE dataFootnote 1, the task of local tumour detection is related to the weakly-supervised object detection (WSOD) that discovers the latent region of interests (ROIs) by only image-level labels. Unlike the WSOD tasks in general computer vision, the global labels of medical data may not cover all latent objects in the image. As shown in Fig. 1(c), the final diagnosis is ‘malignant’, which is determined by a small portion of the local regions while the rest are ‘normal’ tissues that are not included in the global label. In this paper, this observation is called ‘semantic exclusivity’ which leads to another critical task of discriminative feature learning for pCLE data to discover all latent objects in the pCLE video.
To this end, a weakly-supervised feature learning framework (WSFL) is proposed. The architecture of WSFL is illustrated in Fig. 2. Given a set of consecutive pCLE frames, WSFL firstly passes the frames through several convolutional layers which are then processed by fully connected layers to output fixed-size frame-based features. These frame-based features are branched into two different streams: one jointly learns the global-image representation and global diagnosis and the other further learns the frame-based annotations by label propagation. Only global diagnosis labels are used as supervisions to train the two streams based on composite loss. We validate the performance of the representation on dataset with 45 patient cases consisting of 700 pCLE videos. The experiments demonstrate that the proposed method is effective for both global diagnosis and local tumour detection compared to frame-based methods [3, 5] and mosaic-based methods [1, 4].
2 Methodology
2.1 Frame-Based Feature Representation
In this paper, the pCLE data are denoted by \(\{X_i\},i=1,\ldots ,n\) where n is the number of pCLE videos. Each data sample \(X_i\) is composed with \(m_i\) frames \(\{X_{i,j}\}\) where \(j=1,\ldots ,m_i\). The goal of this paper is to learn global prediction function \(H^{g}(\cdot )\) for global diagnosis \(Y^{g}_i\) and local prediction function \(H^{l}(\cdot )\) for local labels \(Y^{l}_{i,j}\) only with the global supervision. Given a pCLE video \(X_i=\{X_{i,j}\},j=1,\ldots ,m_i\), we firstly extract the frame-based features by convolutional neural networks. As shown in Fig. 2, the j-th pCLE frame \(X_{i,j}\) is fed into the convolutional layers and then transformed into a D-dimensional representation \(f^{l}_{i,j}\).
2.2 Local Label Classification
Unlike the frame-based approach, the frame-wise labels are not available during the training procedure in our work. Therefore, one challenge of the proposed method is to infer the labels of all frames based on global diagnosis results. However, it is common that the global label may not cover all regions of the image. This is called ‘semantic exclusivity’ in this paper. As shown in Fig. 1, if the pCLE video/mosaic is annotated with ‘normal’, all its frames should be labeled as ‘normal’. For the ‘benign’ videos, the only confirmed issue is that the video includes at least one frame that indicates the existence of benign regions. Therefore, the status of frames can be either ‘normal’ or ‘benign’. Similarly, the malignant videos are also likely to include benign and normal frames.
In order to infer the labels of all image frames, we built a frame-link graph and propagate the labels between the samples. For all frames \(X_{i,j}\) from the training dataset, the frame-link graph \(G=\{V,E\}\) is constructed where the nodes V are composed with pCLE frames and the edges E with the weight matrix W indicate the similarity between pCLE frames. In this paper, we use the k-NN graph based on RBF-Kernel where the similarity between \(X_{i,j}\) and \(X_{i',j'}\) is calculated by \(\text {exp}(-\Vert f^{l}_{i,j}-f^{l}_{i',j'}\Vert ^2_2/\sigma )\) where \(f^{l}_{i,j}\) and \(f^{l}_{i',j'}\) are the frame-based features. In order to recover the latent ‘normal’ frames in ‘benign’ and ‘malignant’ videos, the labels of frames from normal videos are propagated via the frame-link graph as follows:
where L is the graph Laplacian of the similarity matrix W. Y is the set of labels of all frames after propagation. \(\Vert Y_{normal}-Y_{normal}^*\Vert _2^2\) indicates the frames from normal videos should always be labeled with ‘normal’. The probability of a specific frame belongs to the normal class can be obtained via the label propagation from normal frames. However, the propagation scheme in Eq. (1) has no constraints on global labels. For a benign video, there is at least one frame belonging to the benign class. Therefore, we add the constraints to Eq. (1) as follows:
where \(Y^{l}_i=\{Y^{l}_{i,1},\ldots ,Y^{l}_{i,m_i}\}\) is the vector of labels of pCLE video \(X_i\) and \(\mathbf {1}\) is a all-one vector which has the same number of element with \(Y^{l}_i\); By adding this constraint, the frames which are not likely to be normal are assigned with low confidence for normal class. If the probability is lower than a pre-defined \(\theta \), it can be considered as a benign frame. Similarly, the label propagation is also conducted from the benign videos to malignant videos to recover the latent benign regions. The problem in Eq. (2) can be solved by Augmented Lagrangian method (ALM) [7]. After the label propagation, all frames \(X_{i,j}\) are assigned with the pseudo labels \(\bar{Y}^{l}_{i,j}\). The frame-based classification layers \(H^{l}\) are then trained by minimizing the cross-entropy loss as follows:
2.3 Global Label Classification
The mosaic-based methods take the holistic image as input to generate the global diagnosis result. However, the freehand capture of pCLE data can introduce irregular background in the mosaic image, thus leading to overfitting. Instead of using holistic features of the whole pCLE mosaics, we extract the features for all pCLE frames \(f_{i,j}\) to generate the global features as follows:
where \(\mathcal {F}\) is the max-pooling function. Therefore, the mosaic-based classification layers \(H^{g}\) are trained by minimizing the loss defined as follows:
2.4 Semantic Exclusivity Loss
Although Eqs. (3) and (5) are introduced for global and local classification, the learning of two streams is still separated where only the lower feature extraction layers are shared. In order to preserve the consistency between the global and local results, we introduce the semantic exclusivity loss based on the exclusivity relationship between labels: If the global label is ‘normal’, the ‘benign’ and ‘malignant’ labels are not likely to co-exist in local label sets; If the global label is ‘benign’, there will be no ‘malignant’ local frames. Therefore, the semantic exclusivity loss is defined as follows:
where \(\bar{Y}^{l}\) is the max-pooled label over all frames; \(Y^{g*}\) is the ground-truth of global label where \(Y^{g*}_{i,n}\), \(Y^{g*}_{i,b}\) and \(Y^{g*}_{i,m}\) are the probability of ‘normal’, ‘benign’ and ‘malignant’ respectively. The semantic exclusivity loss can be regarded as an alternative of the standard cross-entropy loss with additional penalizations on the impossible co-existence of local labels.
2.5 Final Objective and Alternative Learning
The final objective is a combination of global classification, local detection and semantic exclusivity loss as follows:
where \(\lambda _{global},\lambda _{local}\) and \(\lambda _{ex}\) are balance weights of each terms. We set \(\lambda _{global}=1\), \(\lambda _{local}=\lambda _{ex}=0.001\) in this paper. It is worth nothing the label propagation process cannot be directly solved via back-propagation. In each epoch, the label propagation is firstly conducted to obtain the pseudo labels for each frame. Then the deep neural networks are trained via back propagation.
3 Experiments
Dataset and Experimental Settings: The dataset is collected by a pre-clinical pCLE system (Cellvizio, Mauna Kea Technologies, Paris, France) as described in [8]. Breast tissue samples are obtained from 45 patients that are diagnosed with three classes including normal, benign and malignant. We finally obtained 700 pCLE mosaics which consist of 8000 frames in total. Among them, 500 pCLE mosaics are used for training and the rest are for testing. For data annotation, each frame is manually labeled with the corresponding class by experts and the mosaics are also labeled with the final diagnosis.
The feature extraction layers in Fig. 2 is based on the residual architecture proposed in [9]. We use the Adam solver [10] with a batch size of 1. The PytorchFootnote 2 framework is adopted to implement the deep convolution neural networks and the experiment platform is a workstation with Xeon E5-2630 and NVIDIA GeForce Titan Xp.
Qualitative Performance Evaluation. We firstly present two typical cases in Fig. 3. The first column is the original pCLE videoFootnote 3; The second column selects local prediction for the representative frames where the green rectangles indicates the normal frames while the red rectangle indicates the malignant frames. Given a new pCLE video, the local and global prediction are updated along with the time frames. For cases 1, several frames at the beginning include the stroma tissues which are similar to the malignant cases. Therefore, the probability of malignant class on both local and global prediction are over 0.1. After receiving sufficient numbers of pCLE frames, the prediction tends to be stable. For case 2, the pCLE starts with normal frames which supports both local and global prediction to ‘normal’. However, the malignant frames exist from frame # 10 to #20 which leads to the global prediction to be ‘malignant’. Although several frames are likely to be normal after frame #20, the global prediction is not changed which demonstrates the proposed method is able to handle the pCLE video with different classes of local cases. More examples can be found in supplementary materials.
Quantitative Performance Evaluation. We also present the quantitative results of global and local prediction on pCLE dataset. The average precision of each class and the mean average precision over all classes are reported to measure the accuracy of classification. Several baselines are implemented in this paper for comparison including dense-SIFT in [11], MVMME in [3], UMGM in [4], Residual CNN [9] and Patch-CNN in [5]. During the model training, all global and local labels are available for baselines while the proposed method is trained with only global supervision. Table 1 shows the classification performance of multiple baseline methods and the proposed WSFL. In overall view, the proposed WSFL achieves the competitive accuracy on both global and local prediction compared to all baselines. For global prediction task, the proposed method outperforms the methods with hand-crafted features even MVMME and UMGM adapt the knowledge from histopathological slides which demonstrated the good feature extraction of convolutional neural networks. However, the CNN model which directly takes the whole mosaic as input does not perform well on global prediction tasks. The main reason is that the pCLE mosaics are resized into the same sizes which are different from the original scales. Compared to the Patch-CNN method, the proposed method recovers the local label based on semantic propagation that helps to learn class-specific features. Moreover, the semantic exclusivity loss further improves the proposed method. For local prediction tasks, the proposed method outperforms most of the baselines even with only global supervision which is also closed to the Patch-CNN trained with frame labels.
4 Conclusion
In this paper, we have proposed a weakly-supervised feature learning (WSFL) framework to learn discriminative features for endomicroscopy analysis. A two-stream convolutional neural networks is adopted to jointly learn global and local prediction based on label propagation and semantic exclusivity loss. Compared to previous frame-based and mosaic-based methods, the proposed framework is trained under the global supervision only while the classification accuracy on both local and global tasks is promising on the breast tissue dataset with 700 pCLE samples. Our future work will focus on reformulating the label propagation process as forward/background operations in neural networks for end-to-end discriminative feature learning.
Notes
- 1.
pCLE videos refer to a set of consecutive frames of pCLE images; pCLE mosaics refer to the image with large field of view which are generated by stitching the frames.
- 2.
- 3.
For better visualization, we present the pCLE mosaics in the experiment. However, the proposed method takes frames as input without mosaicking.
References
André, B., Vercauteren, T., Buchner, A.M., Wallace, M.B., Ayache, N.: A smart atlas for endomicroscopy using automated video retrieval. MedIA 15(4), 460–476 (2011)
Kamen, A., et al.: Automatic tissue differentiation based on confocal endomicroscopic images for intraoperative guidance in neurosurgery. In: BioMed Research International 2016 (2016)
Gu, Y., Yang, J., Yang, G.Z.: Multi-view multi-modal feature embedding for endomicroscopy mosaic classification. In: CVPR, pp. 11–19 (2016)
Gu, Y., Vyas, K., Yang, J., Yang, G.-Z.: Unsupervised feature learning for endomicroscopy image retrieval. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017. LNCS, vol. 10435, pp. 64–71. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66179-7_8
Aubreville, M., et al.: Automatic classification of cancerous tissue in laserendomicroscopy images of the oral cavity using deep learning. Sci. Rep. 7(1), 11979 (2017)
André, B., Vercauteren, T., Buchner, A.M., Wallace, M.B., Ayache, N.: Endomicroscopic video retrieval using mosaicing and visual words. In: IEEE ISBI 2010, pp. 1419–1422. IEEE (2010)
Fortin, M., Glowinski, R.: Augmented Lagrangian Methods: Applications to the Numerical Solution of Boundary-Value Problems, vol. 15. Elsevier, New York (2000)
Chang, T.P., et al.: Imaging breast cancer morphology using probe-based confocal laser endomicroscopy: towards a real-time intraoperative imaging tool for cavity scanning. Breast Cancer Res. Treat. 153(2), 299–310 (2015)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: CVPR, pp. 770–778 (2016)
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980 (2014)
André, B., Vercauteren, T., Perchant, A., Buchner, A.M., Wallace, M.B., Ayache, N.: Endomicroscopic image retrieval and classification using invariant visual features. In: IEEE ISBI 2009, pp. 346–349. IEEE (2009)
Acknowledgement
This research is partly supported by Committee of Science and Technology, Shanghai, China (No. 17JC1403000) and 973 Plan, China (No. 2015CB856004). Yun Gu is supported by Chinese Scholarship Council (CSC). We also thank NVIDIA to provide the device for our work. The tissue specimens were obtained using the Imperial tissue bank ethical protocol following the R-12047 project.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Gu, Y., Vyas, K., Yang, J., Yang, GZ. (2018). Weakly Supervised Representation Learning for Endomicroscopy Image Analysis. In: Frangi, A., Schnabel, J., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science(), vol 11071. Springer, Cham. https://doi.org/10.1007/978-3-030-00934-2_37
Download citation
DOI: https://doi.org/10.1007/978-3-030-00934-2_37
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00933-5
Online ISBN: 978-3-030-00934-2
eBook Packages: Computer ScienceComputer Science (R0)