Abstract
For image segmentation, typical fully convolutional networks (FCNs) need strong supervision through a large sample of high-quality dense segmentations, entailing high costs in expert-raters’ time and effort. We propose MS-Net, a new FCN to significantly reduce supervision cost, and improve performance, by coupling strong supervision with weak supervision through low-cost input in the form of bounding boxes and landmarks. Our MS-Net enables instance-level segmentation at high spatial resolution, with feature extraction using dilated convolutions. We propose a new loss function using bootstrapped Dice overlap for precise segmentation. Results on large datasets show that MS-Net segments more accurately at reduced supervision costs, compared to the state of the art.
Suyash P. Awate—Supported by: Nvidia GPU Grant Program, IIT Bombay Seed Grant 14IRCCSG010, Wadhwani Research Centre for Bioengineering (WRCB) IIT Bombay, Department of Biotechnology (DBT) Govt. of India BT/INF/22/SP23026/2017, iNDx Technology.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
- Instance-level image segmentation
- Fully convolutional networks
- Weak supervision
- Full resolution
- Dice loss
- Bootstrapped loss
1 Introduction and Related Work
Fully convolutional networks (FCNs) are important for segmentation through their ability to learn multiscale per-pixel features. Unlike FCNs for natural-image analysis, FCNs for medical image segmentation cannot always rely on transfer learning of parameters from networks (pre-)trained for natural-image analysis (VGG-16, ResNet). Thus, for medical image segmentation, training FCNs typically needs strong supervision through a large number of high-quality dense segmentations, with per-pixel labels, produced by radiologists or pathologists. However, generating high-quality segmentations is laborious and expensive. We propose a novel FCN, namely, MS-Net, to significantly reduce the cost (time and effort) of expert supervision, and significantly improve performance, by effectively enabling both high-quality/ strong and lower-quality/ weak supervision using training data comprising (i) low-cost coarse-level annotations for a majority of images and (ii) high-quality per-pixel labels for a minority of images.
Early convolutional neural networks (CNNs) for microscopy image segmentation [2] learn features from image patches to label the center-pixel in each patch. Later CNNs [1] use an autoencoder design to extract features from entire brain volumes for lesion segmentation. U-Net [11] localizes objects better by extending the symmetric-autoencoder design to combine high-resolution features from the encoding path with upsampled outputs in the decoding path. Also, U-Net training gives larger weights to misclassification at pre-computed pixel locations heuristically estimated to be close to object boundaries. Similarly, DCAN [4] explicitly adds an additional branch in its FCN to predict the pixel locations close to true object contours. V-Net [9] eliminates U-Net’s heuristic weighting scheme through a loss function based on the Dice similarity coefficient (DSC) to handle a severe imbalance between the number of foreground and background voxels. These segmentation methods lead to reduced precision near object boundaries because of limited context (patches) [2], tiling [11], or subsampling [9]. All these methods rely solely on strong supervision via high-quality, but high-cost, dense segmentations. In contrast, our MS-Net also leverages weak supervision through low-cost input in the form of bounding boxes and landmarks. We improve V-Net’s scheme of using DSC by continuously refocusing the learning on a subset of pixels with predicted class probabilities farthest from their true labels.
Instance segmentation methods like Mask R-CNN [5] simultaneously detect (via bounding boxes) and segment (via per-pixel labels) object instances. Mask R-CNN and other architectures [1, 2, 9, 11] cannot preserve full spatial resolution in their feature maps, and are imprecise in localizing object boundaries. For segmenting street scenes, FRRN [10] combines multiscale context and pixel-level localization using two processing streams: one at full spatial resolution to precisely localize object boundaries and another for sequential feature-extraction and pooling to produce an embedding for accurate recognition. We improve over FRRN by leveraging (i) low-cost weak supervision through bounding-boxes and landmarks, (ii) a bootstrapped Dice (BADICE) based loss, and (iii) dilated convolutions to efficiently use larger spatial context for feature extraction.
We propose a novel FCN architecture for instance-level image segmentation at full resolution. We reduce the cost of expert supervision, and improve performance, by effectively coupling (i) strong supervision through dense segmentations with (ii) weak supervision through low-cost input via bounding boxes and landmarks. We propose the BADICE loss function using bootstrapped DSC, with feature extraction using dilated convolutions, geared for segmentation. Results on large openly available medical datasets show that our MS-Net segments more accurately with reduced supervision cost, compared to the state of the art.
2 Methods
We describe our MS-Net FCN incorporating (i) mixed supervision via dense segmentations, bounding boxes, and landmarks, and (ii) the BADICE loss function.
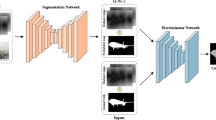
Our MS-Net: Mixed-Supervision FCN for Full-Resolution Segmentation (abstract structure). We enable mixed supervision through a combination of:(i) high-quality strong supervision in the form of dense segmentation (per-pixel label) images, and (ii) low-cost weak supervision in the form of bounding boxes and landmarks. N \(\times \) conv-KxK-D-(S)-[P] denotes: N sequential convolutional layers with kernels of spatial extent (K, K), dilation factor D, spatial stride S, and P output feature maps.
Architecture. Our MS-Net architecture (abstract structure in Fig. 1) has two types of components: (i) a base network for full-resolution feature extraction related to the FRRN [10] architecture and (ii) 3 task-specific subnetwork extensions: segmentation unit (SU), landmark unit (LU), and detection unit (DU).
The base network comprises two streams: (i) the full-resolution residual stream to determine precise object boundaries and (ii) the pooling stream to produce multiscale, robust, and discriminative features. The pooling stream comprises two main components: (i) the residual unit (RU) used in residual networks [6] and (ii) the dilated full-resolution residual unit (DRRU). The DRRU (Fig. 1) takes in two incoming streams and has an associated dilation factor. Features from each stream are first concatenated and then passed through two 3 \(\times \) 3 dilated-convolutional layers, each followed by batch normalization and rectified linear unit (ReLU) activation. The resulting feature map, besides being passed on to the next DRRU, also serves as residual feedback to the full-resolution residual stream afterundergoing channel adjustment using a 1 \(\times \) 1 convolutional layer and subsequent bilinear upsampling. We modify FRRN’s B model (Table 1 in [10]) replacing their 9 groups of full-resolution residual units with an equal number of DRRUs with dilation factors of [1, 1, 2, 2, 4, 2, 2, 1]. The dilated convolutions lend our MS-Net features a larger spatial context to prevent segmentation errors like (i) holes within object regions, where local statistics are closer to the background, and (ii) poor segmentations near image boundaries.
Subnetwork extensions use the features extracted by the base network. The SU takes the extracted features into a 1 \(\times \) 1 convolutional layer followed by a channel-wise softmax to output a full-resolution dense segmentation map.
The LU helps locate landmarks at object boundaries (in this paper) or within objects (in principle). Because LU’s output is closely related SU’s output, we design LU’s input to be identical to SU’s input. LU outputs L mask images, each indicating the spatial location (probabilistically) of one of the L landmarks. To do so, the extracted features are fed through four 3 \(\times \) 3 convolutional layers with a spatial stride of 2. The resulting feature map is fed through a 1 \(\times \) 1 convolutional layer with L output channels to obtain the landmark feature maps at (1/16)-th of the full image resolution. The pixel with the highest activation in the l-th feature map corresponds to the spatial location of the l-th landmark of interest.
Each DU uses DRRU features from different levels in the upsampling path of the pooling stream, to produce object locations, via bounding boxes, and their class predictions for C target classes. Each ground-truth box is represented by (i) a one-hot C-length vector indicating the true class and (ii) a 4-length vector parametrizing the true bounding-box coordinates. A DU uses a single-stage object-detection paradigm, similar to that used in [7]. For each level, at each pixel, the DU outputs \(A := 9\) candidate bounding boxes, termed anchor boxes [7], as follows. The DU’s class-prediction (respectively, location-prediction) subnetwork outputs a C-class probability vector (respectively, 4-length vector) for each of the A anchors. So, the DU passes a DRRU’s T-channel output through four 3 \(\times \) 3 convolutional layers, each with 256 filters, and a 3 \(\times \) 3 convolutional layer with CA (respectively, 4A) filters. To define the DU loss, we consider a subset of anchor boxes that are close to some ground-truth bounding box, with a Jaccard similarity coefficient (JSC) (same as intersection-over-union) >0.5. MS-Net training seeks to make this subset of anchor boxes close to their closest ground-truth bounding boxes. DUs share parameters when their inputs have the number of channels. We pool the class predictions and location predictions from DUs at all levels to get output Z (Fig. 1). During testing, Z indicates a final set of bounding boxes after thresholding the class probabilities.
Loss Functions for SU, LU, DU. Correct segmentations for some image regions are easy to get, e.g., regions far from object and image boundaries or regions without image artifacts. To get high-quality segmentations, training should focus more on the remaining pixels that are hard to segment. U-net [11] and DCAN [4] restrict focus to a subset of hard-to-segment pixels only at object boundaries, thereby failing to capture other hard-to-segment regions, e.g., near image boundaries and image artifacts. In contrast, we use bootstrapped loss, as in [12], by automatically identifying hard-to-segment pixels as the top K percentile with predicted class probabilities farthest from the ground truth; K is a free parameter; typically \(K \in \) [3, 9]. For the SU, our BADICE loss is the mean, over C classes, negative DSC over the top-K pixel subset, where we use the differentiable-DSC between N-pixel probability maps P and Q as \(2 \sum _{n=1}^N P_n Q_n / (\sum _{n=1}^N P_n^2 + \sum _{n=1}^N Q_n^2)\). Indeed, the pixels selected by BADICE (Fig. 2) are near object boundaries as well as other hard-to-segment areas. We find that BADICE leads to faster convergence because the loss-function gradients focus on errors at hard-to-segment pixels and are more informative.
For the LU, the loss function, for the l-th landmark, is the cross-entropy between (i) the binary ground-truth mask (having a single non-zero pixel corresponding to the l-th landmark location) and (ii) a 2D probability map generated by a softmax over all pixels in the l-th channel of the LU output. The DU loss is the mean, over valid anchors, of the sum of (i) a cross-entropy based focal loss [7] on class predictions and (ii) a regularized-\(L_1\) loss for bounding box coordinates.
Training. We minimize the sum of SU, LU, and DU losses using stochastic gradient descent, using checkpoint-based memory optimizations to process memory-intensive dilated convolutions at full-resolution. We use data augmentation through (i) random image resizing by factors \(\in [0.85, 1.25]\) for all datasets and (ii) horizontal and vertical flipping, rotations within \([-25,25]\) degrees, and elastic deformations for histopathology and microscopy data.
3 Results and Discussion
We evaluate 5 methods (free parameters tuned by cross-validation): (i) MS-Net with strong supervision only, via dense segmentation maps; (ii) MS-Net with strong supervision and weak supervision via bounding boxes only; (iii) MS-Net with strong supervision and weak supervision via bounding boxes and landmarks; (iv) U-Net [11]; (v) DCAN [4]. We evaluate all methods at different levels of strong supervision during training, where a fraction of the images have strong-supervision data and the rest have only weak-supervision data. We evaluate on 5 openly available medical datasets. We measure performance by the mean JSC (mJSC), over all classes, between the estimated and true label maps.
Radiographs: Chest. This dataset (Fig. 3(a)–(b)) has 247 high-resolution (2048\(^2\)) chest radiographs (db.jsrt.or.jp/eng.php), with expert segmentations and 166 landmark annotations for 5 anatomical structures (2 lungs, 2 clavicles, heart) [3]. We use the 50-50 training-testing split prescribed by [3]. Qualitatively (Fig. 3(c)–(e)), MS-Net trained with mixed supervision (Fig. 3(c)), i.e., strong supervision via dense label maps and weak supervision via bounding boxes and landmarks, gives segmentations that are much more precise near object boundaries compared to U-net (Fig. 3(d)) and DCAN (Fig. 3(e)) both trained using strong supervision only. Quantitatively (Fig. 4), at all levels of strong supervision, (i) all 3 versions of MS-Net outperform U-net and DCAN, and (ii) MS-Net trained with mixed supervision outperforms MS-Net trained without weak supervision using landmarks or bounding boxes.
Histopathology: Gland. This dataset (Fig. 5(a)–(b)) has 85 training slides (37 benign, 48 malignant) and 80 testing slides (37 benign, 43 malignant) of intestinal glands in human colorectal cancer tissues (warwick.ac.uk/fac/sci/dcs/research/tia/glascontest) with dense segmentations. To create weak-supervision, we generate bounding boxes, but cannot easily generate landmarks. For this dataset, we use DSC and Hausdorff distance (HD) for evaluation, because other methods did this in glascontest. Qualitatively, compared to U-net and DCAN, our MS-Net produces segmentations with fewer false positives (Fig. 5(c)–(e); top left) and better labelling near gaps between spatially adjacent glands (Fig. 5(c)–(e); mid right). Quantitatively, at all strong-supervision levels, (i) MS-Net outperforms U-net and DCAN, and (ii) MS-Net trained with mixed supervision outperforms MS-Net without any weak supervision (Fig. 6).
Microscopy: Cells. mJSC using all training data (strong + weak supervision) for: (a) fluoroscopy, (b) phase-contrast, and (c) differential interference contrast datasets. Box plots give variability over stochasticity in the optimization and train-test splits. (d)–(f) mJSC with different levels of strong supervision for the same 3 datasets.
Microscopy: Cells. The next three datasets [8] (Fig. 7) have cell images acquired using 3 microscopy techniques: (i) fluorescent counterstaining: 43 images, (ii) phase contrast: 35 images, and (iii) differential interference contrast: 20 images. To evaluate weak-supervision, we generate bounding boxes, but cannot easily generate landmarks. We use a random 60-40% training-testing split. Similar to previous datasets, at all strong-supervision levels, our MS-Net outperforms U-net and DCAN qualitatively (Fig. 7) and quantitatively (Fig. 8). U-net and DCAN produces labels maps with holes within cell regions that appear to be similar to the background (Fig. 7(d)-(e)), while our MS-Net (Fig. 7(c)) avoids such errors via BADICE loss and larger-context features through dilated convolutions for multiscale regularity. MS-Net also clearly achieves better boundary localization (Fig. 7(h)), unlike U-net and DCAN that fail to preserve gaps between objects (loss of precision) (Fig. 7(i)–(j)).
Conclusion. For full-resolution segmentation, we propose MS-Net that significantly improves segmentation accuracy and precision, and significantly reduces supervision cost, by effectively coupling (i) strong supervision with (ii) weak supervision through low-cost rater input in the form of bounding boxes and landmarks. We propose (i) BADICE loss using bootstrapped DSC to automatically focus learning on hard-to-segment regions and (ii) dilated convolutions for larger-context features. Results on 5 large medical open datasets clearly show MS-Net’s better performance, even at reduced supervision costs, over the state of the art.
References
Brosch, T., Yoo, Y., Tang, L.Y.W., Li, D.K.B., Traboulsee, A., Tam, R.: Deep convolutional encoder networks for multiple sclerosis lesion segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 3–11. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_1
Ciresan, D., Giusti, A., Schmidhuber, J.: Deep neural networks segment neuronal membranes in electron microscopy images. In: Advances in Neural Information Processing System, pp. 2843–2851 (2012)
Ginneken, B., Stegmann, M., Loog, M.: Segmentation of anatomical structures in chest radiographs using supervised methods: a comparative study on a public database. Med. Image Anal. 10(1), 19–40 (2006)
Hao, C., Xiaojuan, Q., Lequan, Y., Pheng-Ann, H.: DCAN: deep contour-aware networks for accurate gland segmentation. In: IEEE Computer Vision Pattern Recognition, pp. 2487–2496 (2016)
He, K., Gkioxari, G., Dollár, P., Girshick, R.: Mask R-CNN. In: International Conference on Computer Vision, pp. 2980–2988 (2017)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: IEEE Computer Vision Pattern Recognition, pp. 770–778 (2016)
Lin, T., Goyal, P., Girshick, R., He, K., Dollár, P.: Focal loss for dense object detection. In: Intelligence Conference on Computer Vision (2017)
Martin, M., et al.: A benchmark for comparison of cell tracking algorithms. Bioinformatics 30(11), 1609–1617 (2014)
Milletari, F., Navab, N., Ahmadi, S.: V-net: Fully convolutional neural networks for volumetric medical image segmentation. In: International Conference on 3D Vision, pp. 565–571 (2016)
Pohlen, P., Hermans, A., Mathias, M., Leibe, B.: Full-resolution residual networks for semantic segmentation in street scenes. In: IEEE Computer Vision Pattern Recognition, pp. 3309–3318 (2017)
Ronneberger, O., Fischer, P., Brox, T.: U-net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Wu, Z., Shen, C., Hengel, A.: Bridging category-level and instance-level semantic image segmentation (2016). arXiv preprint arXiv:1605.06885
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Shah, M.P., Merchant, S.N., Awate, S.P. (2018). MS-Net: Mixed-Supervision Fully-Convolutional Networks for Full-Resolution Segmentation. In: Frangi, A., Schnabel, J., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science(), vol 11073. Springer, Cham. https://doi.org/10.1007/978-3-030-00937-3_44
Download citation
DOI: https://doi.org/10.1007/978-3-030-00937-3_44
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00936-6
Online ISBN: 978-3-030-00937-3
eBook Packages: Computer ScienceComputer Science (R0)