Abstract
Deep learning approaches to breast cancer detection in mammograms have recently shown promising results. However, such models are constrained by the limited size of publicly available mammography datasets, in large part due to privacy concerns and the high cost of generating expert annotations. Limited dataset size is further exacerbated by substantial class imbalance since “normal” images dramatically outnumber those with findings. Given the rapid progress of generative models in synthesizing realistic images, and the known effectiveness of simple data augmentation techniques (e.g. horizontal flipping), we ask if it is possible to synthetically augment mammogram datasets using generative adversarial networks (GANs). We train a class-conditional GAN to perform contextual in-filling, which we then use to synthesize lesions onto healthy screening mammograms. First, we show that GANs are capable of generating high-resolution synthetic mammogram patches. Next, we experimentally evaluate using the augmented dataset to improve breast cancer classification performance. We observe that a ResNet-50 classifier trained with GAN-augmented training data produces a higher AUROC compared to the same model trained only on traditionally augmented data, demonstrating the potential of our approach.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
1 Introduction
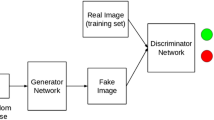
A major enabler of the recent success of deep learning in computer vision has been the availability of massive-scale, labeled training sets (e.g. ImageNet [1]). However, in many medical imaging domains, collecting such datasets is difficult or impossible due to privacy restrictions, the need for expert annotators, and the distribution of data across many sites that cannot share data. The class imbalance naturally present in many medical domains, where “normal” images dramatically outnumber those with findings, further exacerbates these issues (Fig. 1).
Generated samples from ciGAN using previously unseen patches as context. Each row contains (from left to right) the original image, the input to ciGAN, and the synthetic example generated for the opposite class. The first two rows contain examples of the GAN synthesizing a non-malignant patch from a malignant lesion. The third and fourth rows are examples of the GAN synthesizing a malignant lesion on a non-malignant patch, using randomly selected segmentations from other malignant patches. We observe that the GAN is able to incorporate contextual information to smooth out borders of the segmentation masks.
A common technique used to combat overfitting is to synthetically increase the size of a dataset through data augmentation, where affine transformations such as flipping or resizing are applied to training images. The success of these simple techniques raises the question of whether one can further augment training sets using more sophisticated methods. One potential avenue could be to synthetically generate new training examples altogether. While generating training samples may seem counterintuitive, rapid progress in designing generative models (particularly generative adversarial networks (GANs) [2,3,4]) to synthesize highly realistic images merits exploration of this proposal. Indeed, GANs have been used for data augmentation in several recent works [5,6,7,8,9], and investigators have applied GANs to medical images such as magnetic resonance (MR) and computed tomography (CT) [10, 11]. Similarly, GANs have been used for data augmentation in liver lesions [12], retinal fundi [13], histopathology [14], and chest x-rays [15].
A particular domain where GANs could be highly effective for data augmentation is cancer detection in mammograms. The localized nature of many tumors in otherwise seemingly normal tissue suggests a straightforward, first-order procedure for data augmentation: sample a location in a normal mammogram and synthesize a lesion in this location. This approach also confers benefits to the generative model, as only a smaller patch of the whole image needs to be augmented. GANs for data augmentation in mammograms is especially promising because of (1) the lack of large-scale public datasets, (2) the small proportion of malignant outcomes in a normal population (\(\sim \)0.5%) [16] and, most importantly, (3) the clinical impact of screening initiatives, with the potential for machine learning to improve quality of care and global population coverage [17].
Here, we take a first step towards harnessing GAN-based data augmentation for increasing cancer classification performance in mammography. First, we demonstrate that our GAN architecture (ciGAN) is able to generate a diverse set of synthetic image patches at a high resolution (256\(\,\times \,\)256 pixels). Second, we provide an empirical study on the effectiveness of GAN-based data augmentation for breast cancer classification. Our results indicate that GAN-based augmentation improves mammogram patch-based classification by 0.014 AUC over the baseline model and 0.009 AUC over traditional augmentation techniques alone.
2 Proposed Approach: Conditional Infilling GAN
GANs are known to suffer from convergence issues, especially with high dimensional images [3, 4, 18, 19]. To address this issue, we construct a GAN using a multi-scale generator architecture trained to infill a segmented area in a target image. First, our generator is based on a cascading refinement network [20], where features are generated at multiple scales before being concatenated to improve stability at high resolutions. Second, rather than requiring the generator to replicate redundant context in a mammography patch, we constrain the generator to infill only the segmented lesion (either a mass or calcification). Finally, we use a conditional GAN structure to share learned features between non-malignant and malignant cases [21].
The ciGAN generator architecture. The inputs consist of four channels (in blue): one context image (where the lesion is replaced with a random noise mask), one lesion mask, and two class channels for indicating a malignant or non-malignant label. Each convolutional block (in green) represents two convolutional layers with an upsampling operation. (Color figure online)
2.1 Architecture
Our conditional infilling GAN architecture (here on referred to as ciGAN) is outlined in Fig. 2. The input is a concatenated stack (in blue) of one grayscale channel with the lesion replaced with uniformly random values between 0 and 1 (the corrupted image), one channel with ones representing the location of the lesion and zeros elsewhere (the mask), and two channels with values as [1, 0] representing the non-malignant class or [0, 1] as the malignant class (the class labels). The input stack is downsampled to 4\(\,\times \,\)4 and passed into the first convolutional block (in green), which contains two convolutional layers with 3\(\,\times \,\)3 kernels and ReLU activation functions. The output of this block is upsampled to twice the current resolution (8\(\,\times \,\)8) and then concatenated with an input stack resized to 8\(\,\times \,\)8 before being passed into the second convolutional block. This process is repeated until a final resolution of 256\(\,\times \,\)256 is obtained. The convolutional layers have 128, 128, 64, 64, 32, 32, and 32 kernels from the first to the last block. We use the nearest neighbors method for upsampling.
The discriminator network has a similar but inverse structure. The input consists of a 256\(\,\times \,\)256 image. This is passed through a convolutional layer with 32 kernels, 3\(\,\times \,\)3 kernel size, and the LeakyReLU [22] activation function, followed by a 2\(\,\times \,\)2 max pooling operation. We apply a total of 5 convolutional layers, doubling the number of kernels each time until the final layer of 512 kernels. This layer is then flattened and passed into a fully connected layer with one unit and a sigmoid activation function.
2.2 Training Details
Patch-Level Training: Given that most lesions are present within a localized area much smaller than the whole breast image (though context & global features may also be important), we focus on generating patches (256\(\,\times \,\)256) containing such lesions. This allows us to more meaningfully measure the effects of GAN-augmented training as opposed to using the whole image. Furthermore, patch-level pre-training has been shown to increase generalization for full images [23,24,25].
The ciGAN model is trained using a combination of the following three loss functions:
Feature Loss: For a feature loss, we utilize the VGG-19 [26] convolutional neural network, pre-trained on the ImageNet dataset. Real and generated images are passed through the network to extract the feature maps at the \(\text {pool}_1\), \(\text {pool}_2\), and \(\text {pool}_3\) layers, where the mean of the absolute errors is taken between the maps. This loss encourages the features of the generator to match the real image at different spatial resolutions and feature complexities. Letting \({\varPhi _i}\) be the collection of layers in \(\varPhi \), the VGG19 network, where \(\varPhi _0\) is the input image, we define VGG loss for the real image R and generated image S as:
Adversarial Loss: We use the adversarial loss formulated in [27], which seeks optimize over the following mini-max game involving generator G and discriminator D:
Where c is the class label, R is a real image, and S is the generated image.
Boundary Loss: To encourage smoothing between the infilled component and the context of a generated image, we introduce a boundary loss, which is the \(L_{1}\) difference between the real and generated image at the boundary:
Where R is the real image, S is the generated image, w is the mask boundary with a Gaussian filter of standard deviation 10 applied, and \(\odot \) is the element-wise product.
Training Details: In our implementation, we alternate between training the generator and discriminator when the loss for either drops below 0.3. We use the Adam [28] optimizer with \(\beta _1 = 0.9\), \(\beta _2 = 0.999\), \(\epsilon = 10^{-8}\), a learning rate of 1e−4, and batch size of 8. To stabilize training, we first pre-train the generator exclusively on feature loss for 10,000 iterations. Then, we train the generator and discriminator on all losses for an additional 100,000 iterations. We weigh each loss with coefficients 1.0, 10.0, and 10000.0 for GAN loss, feature loss, and boundary loss, respectively.
3 Experiments
3.1 DDSM Dataset
The DDSM (Digital Database for Screening Mammography) dataset contains 10,480 total images, with 1,832 (17.5%) malignant cases and 8,648 (82.5%) non-malignant cases. Image patches are labeled as malignant or non-malignant along with the segmentation masks in the dataset. Both calcifications and masses are used and non-malignant patches contain both benign and non-lesion patches.
We apply a 80% training, 10% validation, and 10% testing split on the dataset. To process full resolution images into patches, we take each image (\(\sim \)5500\(\,\times \,\)3000 pixels) and resize to a target range of 1375\(\,\times \,\)750 while ensuring the original aspect ratio is maintained, as described in [23]. For both non-malignant and malignant cases, we generate 100,000 random 256\(\,\times \,\)256 pixel patches and only accept patches that consist of more than 75% breast tissue.
3.2 GAN-Based Data Augmentation
We evaluate the effectiveness of GAN-based data augmentation on the task of cancer detection. We choose the ResNet-50 architecture as our classifier network [29]. We use the Adam optimizer with an initial learning rate of \(10^{-5}\) and \(\beta _1 = 0.9\), \(\beta _2 = 0.999\), \(\epsilon = 10^{-8}\). To achieve better performance, we initialize the classifier with ImageNet weights. For each regime, we train for 10,000 iterations on a batch size of 32 with a 0.9 learning rate decay rate every 2,000 iterations. The GAN is only trained on the training data used for the classifier.
For traditional image data augmentation, we use random rotations up to 30 degrees, horizontal flipping, and rescaling by a factor between 0.75 and 1.25. For augmentation with ciGAN, we double our existing dataset via the following procedure: for each non-malignant image, we generate a malignant lesion onto it using a mask from another malignant lesion. For each malignant patch, we remove the malignant lesion and generate a non-malignant image in its place. In total, we produce 8,648 synthetically generated malignant patches and 1,832 synthetically generated non-malignant patches. We train the classifier by initially training on equal proportions of real and synthetic data. Every 1000 iterations, we increase the relative proportion of real data used by 20%, such that the final iteration is trained on 90% real data. We observe that this regime helps prevent early overfitting and greater generalization for later epochs.
3.3 Results
Table 1 contains the results of three classification experiments. ciGAN, combined with traditional augmentation, achieves an AUC of 0.896. This outperforms the baseline (no augmentation) model by 0.014 AUC (\(p<0.01\), DeLong method [30]) and traditional augmentation model by 0.009 AUC (\(p<0.05\)). Direct comparison of our results with similar works is difficult given that DDSM does not have standardized training/testing splits, but we find that our models compare on par or favorably to other DDSM patch classification efforts [25, 31, 32].
4 Conclusion
Recent efforts for using deep learning for cancer detection in mammograms have yielded promising results. One major limiting factor for continued progress is the scarcity of data, and especially cancer positive exams. Given the success of simple data augmentation techniques and the recent progress in generative adversarial networks (GANs), we ask whether GANs can be used to synthetically increase the size of training data by generating examples of mammogram lesions. We employ a multi-scale class-conditional GAN with mask infilling (ciGAN), and demonstrate that our GAN indeed is able to generate realistic lesions, which improves subsequent classification performance above traditional augmentation techniques. ciGAN addresses critical issues in other GAN architectures, such as training instability and resolution detail. Scarcity of data and class imbalance are common constraints in medical imaging tasks, and we believe our techniques can help address these issues in a variety of settings.
References
Russakovsky, O., et al.: Imagenet large scale visual recognition challenge. IJCV 115(3), 211–252 (2015)
Goodfellow, I.J., et al.: Generative adversarial nets
Gulrajani, I., Ahmed, F., Arjovsky, M., Dumoulin, V., Courville, A.C.: Improved training of Wasserstein GANs. In: NIPS, pp. 5769–5779 (2017)
Berthelot, D., Schumm, T., Metz, L.: Began: boundary equilibrium generative adversarial networks. arXiv preprint arXiv:1703.10717 (2017)
Peng, X., Tang, Z., Yang, F., Feris, R.S., Metaxas, D.: Jointly optimize data augmentation and network training: adversarial data augmentation in human pose estimation. In: CVPR (2018)
Yu, A., Grauman, K.: Semantic jitter: dense supervision for visual comparisons via synthetic images. Technical report (2017)
Wang, X., Shrivastava, A., Gupta, A.: A-fast-RCNN: hard positive generation via adversary for object detection. arXiv, vol. 2 (2017)
Wang, Y.-X., Girshick, R., Hebert, M., Hariharan, B.: Low-shot learning from imaginary data. arXiv preprint arXiv:1801.05401 (2018)
Antoniou, A., Storkey, A., Edwards, H.: Data augmentation generative adversarial networks. arXiv preprint arXiv:1711.04340 (2017)
Wolterink, J.M., Dinkla, A.M., Savenije, M.H.F., Seevinck, P.R., van den Berg, C.A.T., Išgum, I.: Deep MR to CT synthesis using unpaired data. In: Tsaftaris, S.A., Gooya, A., Frangi, A.F., Prince, J.L. (eds.) SASHIMI 2017. LNCS, vol. 10557, pp. 14–23. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-68127-6_2
Nie, D., et al.: Medical image synthesis with context-aware generative adversarial networks. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017. LNCS, vol. 10435, pp. 417–425. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66179-7_48
Frid-Adar, M., Klang, E., Amitai, M., Goldberger, J., Greenspan, H.: Synthetic data augmentation using GAN for improved liver lesion classification. arXiv preprint arXiv:1801.02385 (2018)
Guibas, J.T., Virdi, T.S., Li, P.S.: Synthetic medical images from dual generative adversarial networks. arXiv preprint arXiv:1709.01872 (2017)
Hou, L., Agarwal, A., Samaras, D., Kurc, T.M., Gupta, R.R., Saltz, J.H.: Unsupervised histopathology image synthesis. arXiv (2017)
Salehinejad, H., Valaee, S., Dowdell, T., Colak, E., Barfett, J.: Generalization of deep neural networks for chest pathology classification in x-rays using generative adversarial networks. arXiv preprint arXiv:1712.01636 (2017)
Cancer.gov: Cancer facts and figures, 2015–2016 (2016)
Ribli, D., Horváth, A., Unger, Z., Pollner, P., Csabai, I.: Detecting and classifying lesions in mammograms with deep learning. Sci. Rep. 8(1), 4165 (2018)
Salimans, T., Goodfellow, I., Zaremba, W., Cheung, V., Radford, A., Chen, X.: Improved techniques for training GANs. In: NIPS, pp. 2234–2242 (2016)
Kodali, N., Abernethy, J., Hays, J., Kira, Z.: How to train your dragan. arXiv preprint arXiv:1705.07215 (2017)
Chen, Q., Koltun, V.: Photographic image synthesis with cascaded refinement networks. In: ICCV 2017, pp. 1520–1529. IEEE (2017)
Mirza, M., Osindero, S.: Conditional generative adversarial nets. arXiv preprint arXiv:1411.1784 (2014)
Xu, B., Wang, N., Chen, T., Li, M.: Empirical evaluation of rectified activations in convolutional network. arXiv (2015)
Lotter, W., Sorensen, G., Cox, D.: A multi-scale CNN and curriculum learning strategy for mammogram classification. In: Cardoso, M.J., et al. (eds.) DLMIA/ML-CDS -2017. LNCS, vol. 10553, pp. 169–177. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-67558-9_20
Nikulin, Y.: DM challenge yaroslav nikulin (therapixel) (2017). Synapse.org
Shen, L.: End-to-end training for whole image breast cancer diagnosis using an all convolutional design. arXiv preprint arXiv:1708.09427 (2017)
Simonyan, K., Zisserman, A.: Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556 (2014)
Goodfellow, I., et al.: Generative adversarial nets. In: NIPS
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980 (2014)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: CVPR, pp. 770–778 (2016)
DeLong, E.R., DeLong, D.M., Clarke-Pearson, D.L.: Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44(3), 837–845 (1988)
Zhu, W., Lou, Q., Vang, Y.S., Xie, X.: Deep multi-instance networks with sparse label assignment for whole mammogram classification. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017. LNCS, vol. 10435, pp. 603–611. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66179-7_69
Lévy, D., Jain, A.: Breast mass classification from mammograms using deep convolutional neural networks. arXiv (2016)
Acknowledgements
This work was supported by the National Science Foundation (NSF IIS 1409097).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Wu, E., Wu, K., Cox, D., Lotter, W. (2018). Conditional Infilling GANs for Data Augmentation in Mammogram Classification. In: Stoyanov, D., et al. Image Analysis for Moving Organ, Breast, and Thoracic Images. RAMBO BIA TIA 2018 2018 2018. Lecture Notes in Computer Science(), vol 11040. Springer, Cham. https://doi.org/10.1007/978-3-030-00946-5_11
Download citation
DOI: https://doi.org/10.1007/978-3-030-00946-5_11
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00945-8
Online ISBN: 978-3-030-00946-5
eBook Packages: Computer ScienceComputer Science (R0)