Abstract
The use of 3D Magnetic Resonance Imaging (MRI) has attracted growing attention for the purpose of diagnosis and treatment planning of intraocular ocular cancers. Precise segmentation of such tumors are highly important to characterize tumors, their progression and to define a treatment plan. Along this line, automatic and effective segmentation of tumors and healthy eye anatomy would be of great value. The major challenge to this end however lies in the disease variability encountered over different populations, often imaged under different acquisition conditions and high heterogeneity of tumor characterization in location, size and appearance. In this work, we consider the Retinoblastoma disease, the most common eye cancer in children. To provide automated segmentations of relevant structures, a multi-sequences MRI dataset of 72 subjects is introduced, collected across different clinical sites with different magnetic fields (3T and 1.5T), with healthy and pathological subjects (children and adults). Using this data, we present a framework to segment both healthy and pathological eye structures. In particular, we make use of a 3D U-net CNN whereby using four encoder and decoder layers to produce conditional probabilities of different eye structures. These are further refined using a Conditional Random Field with Gaussian kernels to maximize label agreement between similar voxels in multi-sequence MRIs. We show experimentally that our approach brings state-of-the-art performances for several relevant eye structures and that these results are promising for use in clinical practice.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
1 Introduction
Retinoblastoma (RB) is the most common form of intraocular cancer with high morbidity and mortality rates in children. To diagnose and treat this cancer, several imaging modalities are typically necessary to properly characterize the tumor, its growth and for any follow-up care. While traditionally 2D Fundus imaging, 2D ultrasound or 3D Computed Tomography (CT) [1, 2] were the modalities of choice, 3D Magnetic Resonance Imaging (MRI) has gained increased interest within the ophthalmic community thanks to the high spatial resolutions, multiplanar capabilities and high intrinsic contrast [4, 5]. In effect, 3D MRI thus allows for clear overall improved discrimination between anatomical structures and different pathological regions such as the gross tumor volume, retinal detachment, and intraocular bleeding as illustrated in Fig. 1. As such, automatic and effective segmentation of tumors and healthy eye anatomy would be of great value for both disease diagnosis and treatment planning. For example, having reliable MR imaging biomarkers would open the door to eye cancer radiogenomics [3] (the association of radiological image features with gene expression profile) which can support in prognosis and patient selection for targeted treatment, thereby contributing to precision medicine.
Towards this goal, previous methods based on geometrical and statistical models had addressed ocular segmentation in 3D medical imaging (e. g., MRI or CT). For instance, a parametric model allowed for coarse eye structure segmentations [6], while [7] introduced a 3D shape model of the retina to study abnormal shape changes and peripheral vision. Similarly, 3D mesh construction with morphologic parameters such as distance from the posterior corneal pole and deviation from sphericity have also been proposed [8], as well as using Active Shape Models (ASM) to analyze eye shape information [9, 10]. Unfortunately, a major limitation of the aforementioned methods is that they focus solely on healthy eyes, while the characterization of tumors it self has by and large not been addressed. Actually, ocular tumor segmentation is challenging because of small amount of data but images acquired under different conditions and huge variability of tumors in location, size and appearance.
To this end, we propose a fully automated framework capable of delineating both healthy structures and RB tumors from multi-contrast 3D MRI. It includes data pre-processing for normalization, multi-sequence MRI registration, an effective coarse segmentation and output post-processing to improve localization accuracy. Our approach is based on the popular UNet Convolutional Neural Network (CNN) architecture [11], whereby we segment different healthy eye structures and the tumor in a single step. From this multi-class 3D segmentation, we further refine our estimate by using a Gaussian edge potential Conditional Random Field (CRF) to maximize label agreement between similar voxels in the multi-sequence MRIs. Although we applied here the original implementations of above methods, their combination and the application context is novel with the possibility to easily be extended to other types of tumors. We compare our proposed framework with state-of-the-art techniques on a large mixed dataset, including both healthy and pathological eyes as well as children and adult data from different magnetic fields and MR sequences. Our method allows simultaneous segmentation of both healthy and tumor regions to be identified and outperforms existing approaches used a mixture of ASM [9, 10] and deep learning [12].
2 Methodology
2.1 Dataset
Originating from two clinical centers, our study contains 72 eyes consisting of 32 RB, 16 healthy children (HC) eyes, and 24 healthy adult (HA) eyes. All MRI examinations were performed with a Siemens scanner (SIEMENS Magnetom Aera, Erlangen, Germany) with both T1-weighted (T1w) and T2-weighted (T2w) contrasts. A 3T MR with a head coil was used to image asleep children aged 4 months to 8 years old (mean age of \(3.29\pm 2.15\) y.o.), with a cohort eye diameter size mean of \(12.9\pm 1.3\) mm (range [10.4–15.9] mm). A 1.5T MR with surface coil was used for awake adults aged \(28.4\pm 5.2\) years old (range [23–46]y.o.), with a cohort eye size mean of \(24.7\pm 0.6\) mm (range [23.3–26] mm). The study was approved by the Ethics Committee of the involved institutions and all subjects provided written informed consent prior to participation. All subject information in our study was anonymized and de-identified. Table 1 shows the different parameters used for the two MRI acquisition protocols. Whenever children and adult images are used together we denote mixed cohort (MC).
MRI Data Normalization: Clinical image quality is affected by many factors such as noise, low varying intensity, variations due to non-uniform magnetic field, imperfections of coils, magnetic susceptibility at interfaces, all of which can be influenced by different imaging parameters such as signal-to-noise ratio or acquisition time. In order to compensate for such effects, all MRI volumes were pre-processed with an anisotropic diffusion filtering [14], to reduce noise without removing significant image content. We applied the N4 algorithm [15] to correct for bias field variations and performed histogram-based intensity normalization [16] to build an intensity profile of the dataset. In order to improve the performance in segmentation and computation time, we defined a volume of interest (VOI) of the eye by retaining a \(72\times 72\times 64\) volume centered on the eye such that the optic nerve was always included. Rigid registration was applied to move T2w images into T1w image space.
Manual Segmentation: For training and validation purposes, manual delineations of the eye lens, sclera, tumors and optic nerve were done by radiation oncologist expert. First, segmentation for sclera and tumor was individually by intensity thresholding. Then, manual editing was done to refine borders and remove outlier regions. For small structures such as the lens and the optic nerve, manual segmentations were performed directly using a stylus.
2.2 Automated Anatomical Structure Segmentation
Coarse segmentation: Similar to the
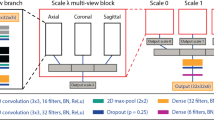
original UNet method presented in [11], we consider an encoder and decoder network that takes as input multiple image channels for each of the imaging sequence types (see Fig. 2). Each encoding and decoding pathway contains 4 layers that effectually changes the feature dimension (i. e., 32, 64, 128, 256, 512). The same architecture accounts for the decoding pathway. In each case, \(3\times 3\times 3\) Convolutions are used with a Batch normalization and parametric rectified linear unit (PRelu) operations.
Between two layers in the encoder pathway, \(2\times 2\times 2\) max pooling with strides of two in each dimension are used. In the decoder pathway, the connection of two subsequent layers is performed with an up-convolution of \(2\times 2\times 2\) with strides of two in each dimension. Concatenation is performed to connect the output tensors of two layers of the encoder and decoder pathways at same level. To train our network, we used the Adam optimizer and the Dice loss function. At inference time, softmax is used to extract probability maps for each class.
3D Fully Connected CRF with Gaussian Kernels: To provide a more refined segmentation, we use a 3D CRF [17] to maximize label agreement between similar voxels (or patches) in the multi-sequences MRI. The 3D CRF incorporates unary potentials of individual voxels and pair-wise potentials (in terms of appearance and smoothness) on neighboring voxels to provide more accurate eye structure segmentation.
Considering an input image I and a probability map P (i. e., provided by the above network), the unary potential is defined to be the negative log-likelihood \(\psi _u(z_i)=-logP(z_i|I)\), where \(z_i\) the predicted label of voxel i. The pair-wise potential has the form \(\psi _p(z_i,z_j)= \mu (z_i,z_j)k(f_i,f_j)\), where \(\mu \) is a label compatibility function, \(k(f_i,f_j)\) and is characterized by integrating two Gaussian kernels of appearance (first term) and smoothness (second term), as follows
where \(p_i\) are voxel locations, \(I_i\) are voxel intensities, \(f_i\) are voxel feature vectors as described in [13], \(w_j\) are weight factor between the two terms, and the \(\theta \)’s are tunable parameters of the Gaussian kernels. The Gibbs energy of CRF model is then given by \(\sum \left( \psi _u(z_i),\psi _p(z_i,z_j)\right) \) [17].
3 Experiments
We performed leave-one-out cross-validations to quantitatively compare the results of the proposed segmentation scheme (i. e., iteratively chose one eye as a validation case, while the remaining subjects are used as the training set). The quality of the segmentations were evaluated by computing the predicted and true volume overlap using the Dice similarity coefficient (DSC) and the Hausdorff distance (HD). For the training step, we crop volumes to size \(72\times 72\times 64\). We report the best performance result obtained from the different parameter settings detailed as follows. For 3D Unet: regularisation type {L1,L2}; number of samples per volume {8, 16, 32}; volume padding size {8, 16, 32}; learning rate for the optimiser {0.001, 0.005}; maximum iterations {0.001, 0.005}. For CRF: neighborhood size {[3,3,3], [5,5,5]}; intensity-homogeneous distance {[5,5,5], [10,10,10]}; kernel weights of appearance and smoothness terms {[1,1], [3,1], [1,3]}. The performance of the proposed method compared to two baselines algorithms found in the literature: an ASM method [9, 10] and a 3D CNN [12].
First, we compare the proposed method with that of an ASM [10] on 40 healthy eyes (24 HA subjects with a 1.5T MRI system and 16HC subjects in a 3T MRI system). Table 2 (top half) reports quality measures and indicates that our approach performs slightly better on the sclera and optic nerve but not on the lens. Indeed, the sclera and the optic nerve have large anatomical variability, due in part to large differences in eye size. The ASM is limited in its ability to take these large variations into account (see Fig. 3 first column, where the smallest healthy eye was used as testing input).
Second, we evaluate the segmentation accuracy of healthy structures in presence of RB tumors (see Table 2 (botom half)). Two training scenarios are considered: (1) 48 children eyes from 3T MR images (i. e., 32RB + 16HC) and, (2) the mixed cohort, MC (i. e., 32RB + 16HC + 24HA) described in Sect. 2.1. Segmentation results on the sclera, lens and optic nerve are superior when using the MC. However, no statistical differences (Wilkoxon signed rank test) were found.
RB segmentation results are presented as function of its size in Fig. 4. The mean DSC and HD using a MC training is of \(59.1\pm 12.4\%\) and \(5.33\pm \)2.54 mm, respectively. When compared with a training set using children eyes only, our approach yields gains of \(1.2\%\) DSC and \(0.21\,\)mm HD (mean DSC of \(57.9\pm 13.2\%\) and mean HD of \(5.54\pm 2.65\) mm). Similarly to healthy structures, these results indicate that our approach benefits from using the MC dataset and that healthy and children eyes, regardless of what scanner used to image them, can be used jointly to improve segmentation performances. Let us note that differences in DSC were statistically significant (\(p<.005\)). Qualitative results are shown in second and third column of Fig. 3. Finally, as regards similar approaches in the literature [12], our method provide slight improvements (reported results in [12] on 16 RB were of the sclera (\(94.62\pm 1.9\%\)), lens (\(85.67\pm 4.68\%\)), optic nerve (absent) and RB tumor (\(62.25\pm 26.27\%\)) for DICE overlap). However, given that their results are achieved using a different and smaller dataset, a direct comparison would not be fair.
4 Conclusion
In this paper, we have explored the problem of simultaneous segmentation of eye structures from multi-sequences MR images to support clinicians in their need of precise tumor characterization and their progression. We proposed a thorough segmentation pipeline consisting of a combination of data quality normalization and a 3D Unet CNN segmentation model with a Gaussian kernel CRF framework. Effectively, the proposed method embeds the probability maps, the output of 3D Unet architecture, with respect to the analysis of pair-wise appearance and smoothness on neighborhood voxels using CRF model. We validated our method with a heterogeneous eye dataset consisting of a diverse population (adults and children) acquired over multiple sites with different MRI acquisition conditions. Differing from state-of-the-art ASM methods, the proposed method offers an accurate and fully automatic segmentation without any prior computations of statistics on the shape of the eye and its structures. Surprisingly, we show here that our method is also largely robust to the eye size and imaging acquisition conditions. Our approach can be easily extended to other types of occular tumors (e. g., Uveal melanoma) to provide an effective and automated support in clinical practice (diagnosis, treatment planning and follow-up).
References
Kook, D., et al.: Variability of standardized echographic ultrasound using 10 mHz and high-resolution 20 mHz B scan in measuring intraocular melanoma. Clin. Ophthal. 5, 477–482 (2011)
Ruegsegger, M.D., et al.: Statistical modeling of the eye for multimodal treatment planning for external beam radiation therapy of intraocular tumors. Int. J. Radiat. Oncol. Biol. Phys. 84(4), 541–547 (2012)
Jansen, R. et al.: MR imaging features of retinoblastoma: association with gene expression profiles. Radiology (2018)
De Graaf, P., et al.: Guidelines for imaging retinoblastoma: imaging principles and MRI standardization. Pediatr. Radiol. 42(1), 2–14 (2014)
Tartaglione, T., et al.: Uveal melanoma: evaluation of extrascleral extension using thin-section MR of the eye with surface coils. La Radio. Med. 119(10), 775–783 (2014)
McCaffery, S., et al.: Three-dimensional high-resolution magnetic resonance imaging of ocular and orbital malignancies. Archiv. Ophthal. 120, 747–754 (2002)
Beenakker, J., et al.: Automated retinal topographic maps measured with magnetic resonance imaging. Invest. Ophthalmol. Vis. Sci. 56, 1033–1039 (2015)
Singh, K., et al.: Three-dimensional modeling of the human eye based on magnetic resonance imaging. Invest. Ophthalmol. Vis. Sci. 47, 2272–2279 (2006)
Ciller, C., et al.: Automatic segmentation of the eye in 3D magnetic resonance imaging a novel statistical shape model for treatment planning of retinoblastoma. Int. J. Radiat. Oncol. Biol. Phys. 92(4), 94–802 (2015)
Nguyen H.-G., et al.: Personalized anatomic eye model from T1-weighted VIBE MR imaging of patients with Uveal melanoma. J. Radiat. Oncol. Biol. Phys. (2018)
Çiçek, Ö., Abdulkadir, A., Lienkamp, S.S., Brox, T., Ronneberger, O.: 3D U-net: learning dense volumetric segmentation from sparse annotation. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016 Part II. LNCS, vol. 9901, pp. 424–432. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_49
Ciller, C., et al.: Multi-channel MRI segmentation of eye structures and tumors using patient-specific features. PLoS ONE 12(3), e173900 (2017)
Kamnitsas, K., et al.: Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 36, 61–78 (2017)
Perona, P., et al.: Scale-space and edge detection using anisotropic diffusion. IEEE Trans. Pattern Anal. Mach. Intell. 12(7), 629–639 (1990)
Tustison, N., et al.: N4ITK: improved N3 bias correction. IEEE Trans. Med. Imaging 29(6), 1310–1320 (2010)
Nyul, L., et al.: New variants of a method of MRI scale standardization. IEEE Trans. Med. Imaging 19(2), 143–50 (2000)
Krähenbühl, P.: Efficient inference in fully connected CRFs with Gaussian edge potentials. Adv. Neural Inf. Process. Syst. 24, 109–117 (2011)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Nguyen, HG. et al. (2018). Ocular Structures Segmentation from Multi-sequences MRI Using 3D Unet with Fully Connected CRFs. In: Stoyanov, D., et al. Computational Pathology and Ophthalmic Medical Image Analysis. OMIA COMPAY 2018 2018. Lecture Notes in Computer Science(), vol 11039. Springer, Cham. https://doi.org/10.1007/978-3-030-00949-6_20
Download citation
DOI: https://doi.org/10.1007/978-3-030-00949-6_20
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00948-9
Online ISBN: 978-3-030-00949-6
eBook Packages: Computer ScienceComputer Science (R0)