Abstract
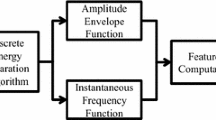
Identifying the features that contribute to classification using machine learning remains a challenging problem in terms of the interpretability and computational complexity of the endeavor. Especially in electroencephalogram (EEG) medical applications, it is important for medical doctors and patients to understand the reason for the classification. In this paper, we thus propose a method to quantify contributions of interpretable EEG features on classification using the Shapley sampling value (SSV). In addition, a pruning method is proposed to reduce the SSV computation cost. The pruning is conducted on an EEG feature tree, specifically at the sensor (electrode) level, frequency-band level, and amplitude-phase level. If the contribution of a feature at a high level (e.g., sensor level) is very small, the contributions of features at a lower level (e.g., frequency-band level) should also be small. The proposed method is verified using two EEG datasets: classification of sleep states, and screening of alcoholics. The results show that the method reduces the SSV computational complexity while maintaining high SSV accuracy. Our method will thus increase the importance of data-driven approaches in EEG analysis.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Ancona, M., Ceolini, E., Oztireli, C., Gross, M.: Towards better understanding of gradient-based attribution methods for deep neural networks. In: Proceedings of the 6th International Conference on Learning Representations (2018)
Asuncion, A., Newman, D.: UCI Machine Learning Repository (2007)
Bach, S., Binder, A., Montavon, G., Klauschen, F., Muller, K.R., Samek, W.: On pixel-wise explanations for non-linear classifier decisions by layer-wise relevance propagation. PLoS ONE 10(7), e0130140 (2015)
Goldberger, A.L., et al.: Physiobank, physiotoolkit, and physionet. Circulation 101(23), e215–e220 (2000)
Hartmann, K.G., Schirrmeister, R.T., Ball, T.: Hierarchical internal representation of spectral features in deep convolutional networks trained for EEG decoding. In: Proceedings of the 6th International Conference on Brain-Computer Interface, pp. 1–6 (2018)
Lawhern, V.J., Solon, A.J., Waytowich, N.R., Gordon, S.M., Hung, C.P., Lance, B.J.: EEGNet: a compact convolutional network for EEG-based brain-computer interfaces. arXiv:1611.08024 (2016)
Li, Y., et al.: Targeting EEG/LFP synchrony with neural nets. In: Advances in Neural Information Processing Systems, pp. 4623–4633 (2017)
Lundberg, S.M., Lee, S.I.: A unified approach to interpreting model predictions. In: Advances in Neural Information Processing Systems, pp. 4768–4777 (2017)
Mumtaz, W., Vuong, P.L., Malik, A.S., Rashid, R.B.A.: A review on EEG-based methods for screening and diagnosing alcohol use disorder. Cogn. Neurodynamics, 1–16 (2018)
Ribeiro, M.T., Singh, S., Guestrin, C.: Why should I trust you? Explaining the predictions of any classifier. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp. 1135–1144. ACM (2016)
Schirrmeister, R.T., et al.: Deep learning with convolutional neural networks for EEG decoding and visualization. Hum. Brain Mapp. 38(11), 5391–5420 (2017)
Shapley, L.S.: A value for n-person games. Contrib. Theory Games 2(28), 307–317 (1953)
Shrikumar, A., Greenside, P., Kundaje, A.: Learning important features through propagating activation differences. arXiv:1704.02685 (2017)
Shtrumbelj, E., Kononenko, I.: Explaining prediction models and individual predictions with feature contributions. Knowl. Inf. Syst. 41(3), 647–665 (2014)
Sturm, I., Lapuschkin, S., Samek, W., Muller, K.R.: Interpretable deep neural networks for single-trial EEG classification. J. Neurosci. Methods 274, 141–145 (2016)
Sundararajan, M., Taly, A., Yan, Q.: Axiomatic attribution for deep networks. arXiv:1703.01365 (2017)
Tcheslavski, G.V., Gonen, F.F.: Alcoholism-related alterations in spectrum, coherence, and phase synchrony of topical electroencephalogram. Comput. Biol. Med. 42(4), 394–401 (2012)
Vilamala, A., Madsen, K.H., Hansen, L.K.: Deep convolutional neural networks for interpretable analysis of EEG sleep stage scoring. arXiv:1710.00633 (2017)
Acknowledgements
This work was supported by the Center of Innovation Program from Japan Science and Technology Agency and JST CREST Grant Number JPMJCR17A4, Japan.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Tachikawa, K., Kawai, Y., Park, J., Asada, M. (2018). Effectively Interpreting Electroencephalogram Classification Using the Shapley Sampling Value to Prune a Feature Tree. In: Kůrková, V., Manolopoulos, Y., Hammer, B., Iliadis, L., Maglogiannis, I. (eds) Artificial Neural Networks and Machine Learning – ICANN 2018. ICANN 2018. Lecture Notes in Computer Science(), vol 11141. Springer, Cham. https://doi.org/10.1007/978-3-030-01424-7_66
Download citation
DOI: https://doi.org/10.1007/978-3-030-01424-7_66
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-01423-0
Online ISBN: 978-3-030-01424-7
eBook Packages: Computer ScienceComputer Science (R0)