Abstract
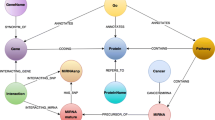
Health and life sciences’ research fields like personalized medicine, drug discovery, pharmacovigilance and systems biology make an intensive use of graphical information to represent knowledge in the form of domain-specific diagrams, such as molecular pathway‘s. The aim is to provide added value to written text in scientific literature and related documents. Enabling access to all the existing literature for further research requires enabling access to the information contained in these diagrams. Molecular pathways are very different from more conventional diagrams (e.g. flowcharts), and therefore interpretation of molecular pathway diagrams requires domain-specific knowledge to remove ambiguity. In this paper, we propose a method that automatically extracts information from molecular pathways using computer vision techniques. To the best of our knowledge this is the first attempt to retrieve information from images depicting molecular pathway diagrams. The lack of a significant, publicly available dataset with annotated ground truth has led to experimental evaluation on synthetic data. Results show high precision and recall values for the detection of entities and relations. We compare and describe the substantial differences between the proposed method and prior art on the closest diagram type using CLEF-IP flowchart summarization task.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Notes
- 1.
https://www.ncbi.nlm.nih.gov/pmc/, as of March 2018.
References
Brocke, J.V., et al.: Reconstructing the giant: On the importance of rigour in documenting the literature search process. In: ECIS Proceedings (2009)
Müller, H., Foncubierta-Rodríguez, A., Lin, C., Eggel, I.: Determining the importance of figures in journal articles to find representative images. In: SPIE Proceedings, vol. 8674 (2013)
Fabregat, A., et al.: The reactome pathway knowledgebase. Nucleic Acids Res. 44(D1), D481–D487 (2016)
Petri, V., et al.: The pathway ontology - updates and applications. J. Biomed. Semant. 5(1), 7 (2014)
Davis, A.P., et al.: The comparative toxicogenomics database: update 2011. Nucleic Acids Res. 39(Database issue), D1067–D1072 (2011)
Hayman, G.T., et al.: The updated RGD pathway portal utilizes increased curation efficiency and provides expanded pathway information. Hum. Genomics 7, 4 (2013)
Paley, S.M., Latendresse, M., Karp, P.D.: Regulatory network operations in the pathway tools software. BMC Bioinformatics 13, 243 (2012)
Ravikumar, K.E., Wagholikar, K.B., Liu, H.: Challenges in adapting text mining for full text articles to assist pathway curation. In: Proceedings of the 5th ACM Conference on Bioinformatics, Computational Biology, and Health Informatics, BCB 2014, pp. 551–558. ACM, New York (2014)
García-Jiménez, B., Pons, T., Sanchis, A., Valencia, A.: Predicting protein relationships to human pathways through a relational learning approach based on simple sequence features. IEEE/ACM Trans. Comput. Biol. Bioinform. 11(4), 753–765 (2014)
Yoon, S., et al.: Systematic identification of context-dependent conflicting information in biological pathways. In: Proceedings of the ACM 8th International Workshop on Data and Text Mining in Bioinformatics, DTMBIO 2014, p. 9. ACM, New York (2014)
Luna, A., Sunshine, M.L., van Iersel, M.P., Aladjem, M.I., Kohn, K.W.: PathVisio-MIM: Pathvisio plugin for creating and editing molecular interaction maps (MIMs). Bioinformatics 27(15), 2165–2166 (2011)
Wang, Y.T., Huang, Y.H., Chen, Y.C., Hsu, C.L., Yang, U.C.: PINT: pathways integration tool. Nucleic Acids Res. 38(Web Server issue), W124–W131 (2010)
Le Novere, N., et al.: The systems biology graphical notation, 27(8) 735–741
Hucka, M., et al.: The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models, 19(4), 524–531
Garcia Seco de Herrera, A., Kalpathy-Cramer, J., Demner-Fushman, D., Antani, S., Müller, H.: Overview of the ImageCLEF 2013 medical tasks. In: CLEF (Working Notes) (2013)
Müller, H., Kalpathy-Cramer, J., Demner-Fushman, D., Antani, S.: Creating a classification of image types in the medical literature for visual categorization. In: Proceedings SPIE 8319, Medical Imaging 2012: Advanced PACS-based Imaging Informatics and Therapeutic Applications, vol. 8319, pp. 83190P–83190P-12 (2012)
Foncubierta-Rodríguez, A., García Seco de Herrerea, A. Müller, H.: Medical image retrieval using bag of meaningful visual words: unsupervised visual vocabulary pruning with PLSA. In: Proceedings of the 1st ACM International Workshop on Multimedia Indexing and Information Retrieval for Healthcare, MIIRH 2013, pp. 75–82. ACM (2013)
Puddu, A., Mach, F., Nencioni, A., Viviani, G.L., Montecucco, F.: An Emerging Role of Glucagon-Like Peptide-1 in Preventing Advanced-Glycation-End-Product-Mediated Damages in Diabetes. Mediators of Inflammation 2013 (2013)
Enders, G.H.: Gauchos and ochos: a Wee1-Cdk tango regulating mitotic entry. Cell Div. 5, 12 (2010)
Kim, H.L., Seo, Y.R.: Molecular and genomic approach for understanding the gene-environment interaction between Nrf2 deficiency and carcinogenic nickel-induced DNA damage. Oncol. Rep. 28(6), 1959–1967 (2012)
Futrelle, R.P.: Strategies for diagram understanding: generalized equivalence, spatial/object pyramids and animate vision. In: Proceedings of the Conference on 10th International Pattern Recognition, vol. 1, pp. 403–408 (1990)
Futrelle, R.P., Kakadiaris, I.A., Alexander, J., Carriero, C.M., Nikolakis, N., Futrelle, J.M.: Understanding diagrams in technical documents. Computer 25(7), 75–78 (1992)
Lank, E., Thorley, J., Chen, S., Blostein, D.: On-line recognition of UML diagrams. In: Proceedings of Sixth International Conference on Document Analysis and Recognition, Institute of Electrical & Electronics Engineers (IEEE) (2001)
Zheng, W.T., Sun, Z.X.: Knowledge-based hierarchical sketch understanding. In: Proceedings of International Conference Machine Learning and Cybernetics, vol. 5, pp. 2838–2843, August 2005
Hammond, T., Davis, R.: Tahuti: a geometrical sketch recognition system for UML class diagrams. In: ACM SIGGRAPH 2006 Courses, SIGGRAPH 2006. ACM, New York (2006)
Thean, A., Deltorn, J.M., Lopez, P., Romary, L.: Textual summarisation of flowcharts in patent drawings for CLEF-IP 2012. In: CLEF 2012 (2012)
Lupu, M., Piroi, F., Hanbury, A.: Evaluating flowchart recognition for patent retrieval. In: EVIA@ NTCIR (2013)
Rusiñol, M., de las Heras, L.P., Terrades, O.R.: Flowchart recognition for non-textual information retrieval in patent search. Inf. Retrieval 17(5-6), 545–562 (2014)
Forbus, K.D., Usher, J., Chapman, V.: Sketching for military courses of action diagrams. In: Proceedings of the 8th International Conference on Intelligent User Interfaces, IUI 2003, pp. 61–68. ACM, New York (2003)
Mas, J., Sanchez, G., Llados, J., Lamiroy, B.: An incremental on-line parsing algorithm for recognizing sketching diagrams. In: Ninth International Conference on Document Analysis and Recognition (ICDAR 2007), Institute of Electrical & Electronics Engineers (IEEE), September 2007
Feng, G., Viard-Gaudin, C., Sun, Z.: On-line hand-drawn electric circuit diagram recognition using 2D dynamic programming. Pattern Recogn. 42(12), 3215–3223 (2009)
Nakamura, Y., Furukawa, R., Nagao, M.: Diagram understanding utilizing natural language text. In: Proceedings of Second International Document Analysis and Recognition Conference, pp. 614–618, October 1993
Butler, G., Grogono, P., Shinghal, R., Tjandra, I.: Retrieving information from data flow diagrams. In: Proceedings of 2nd Working Conference Reverse Engineering, pp. 22–29, July 1995
Watanabe, Y., Nagao, M.: Diagram understanding using integration of layout information and textual information. In: Proceedings of the 17th International Conference on Computational Linguistics, COLING 1998, vol. 2, pp. 1374–1380. Association for Computational Linguistics, Stroudsburg (1998)
Mörzinger, R., Schuster, R., Horti, A., Thallinger, G.: Visual structure analysis of flow charts in patent images. In: CLEF (Online Working Notes/Labs/Workshop) (2012)
Rusiñol, M., et al.: CVC-UAB’s participation in the flowchart recognition task of CLEF-IP 2012. In: CLEF (Online Working Notes/Labs/Workshop) (2012)
Smith, R.: An overview of the Tesseract OCR engine. In: Proceedings of the Ninth International Conference on Document Analysis and Recognition, ICDAR 2007, vol. 02, pp. 629–633. IEEE Computer Society, Washington, DC (2007)
Harris, C., Stephens, M.: A combined corner and edge detector. In: Alvey Vision Conference, vol. 15, p. 50. Citeseer (1988)
Su, Z., Yang, Z., Xu, Y., Chen, Y., Yu, Q.: MicroRNAs in apoptosis, autophagy and necroptosis. Oncotarget 6(11), 8474–8490 (2015)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Foncubierta-Rodríguez, A., Ciubotaru, AN., Bekas, C., Gabrani, M. (2018). Extracting Information from Molecular Pathway Diagrams. In: Fornés, A., Lamiroy, B. (eds) Graphics Recognition. Current Trends and Evolutions. GREC 2017. Lecture Notes in Computer Science(), vol 11009. Springer, Cham. https://doi.org/10.1007/978-3-030-02284-6_8
Download citation
DOI: https://doi.org/10.1007/978-3-030-02284-6_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-02283-9
Online ISBN: 978-3-030-02284-6
eBook Packages: Computer ScienceComputer Science (R0)