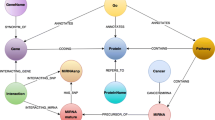
Abstract
Genome analysis is a major precondition for future advances in the life sciences. The complex organization of genome data and the interactions between genomic components can often be modeled and visualized in graph structures. In this paper we propose the integration of several data sets into a graph database. We study the aptness of the database system in terms of analysis and visualization of a genome regulatory network (GRN) by running a benchmark on it. Major advantages of using a database system are the modifiability of the data set, the immediate visualization of query results as well as built-in indexing and caching features.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Albers, D., Dewey, C., Gleicher, M.: Sequence surveyor: leveraging overview for scalable genomic alignment visualization. IEEE Trans. Vis. Comput. Graph. 17(12), 2392–2401 (2011)
van Arensbergen, J., van Steensel, B., Bussemaker, H.J.: In search of the determinants of enhancer-promoter interaction specificity. Trends Cell Biol. 24(11), 695–702 (2014). http://www.sciencedirect.com/science/article/pii/S0962892414001184
Baker, C.A., Carpendale, M.S.T., Prusinkiewicz, P., Surette, M.G.: GeneVis: visualization tools for genetic regulatory network dynamics. In: Visualization, VIS 2002. IEEE. pp. 243–250 (2002)
Bozdag, S., Li, A., Wuchty, S., Fine, H.A.: FastMEDUSA: a parallelized tool to infer gene regulatory networks. Bioinformatics 26(14), 1792–1793 (2010)
Van den Bulcke, T., et al.: SynTReN: a generator of synthetic gene expression data for design and analysis of structure learning algorithms. BMC Bioinform. 7(1), 43 (2006). https://doi.org/10.1186/1471-2105-7-43
Chatraryamontri, A., et al.: The BioGRID interaction database: 2015 update. Nucleic Acids Res. (2014). http://nar.oxfordjournals.org/content/early/2014/11/26/nar.gku1204.abstract
Fiannaca, A., La Rosa, M., La Paglia, L., Messina, A., Urso, A.: BioGraphDB: a new graphDB collecting heterogeneous data for bioinformatics analysis. In: Proceedings of BIOTECHNO (2016)
Gomez, J., et al.: BioJS: an open source Javascript framework for biological data visualization. Bioinformatics 29(8), 1103–1104 (2013). https://doi.org/10.1093/bioinformatics/btt100
Have, C.T., Jensen, L.J.: Are graph databases ready for bioinformatics? Bioinformatics 29(24), 3107 (2013)
Jupiter, D., Chen, H., VanBuren, V.: STARNET2: a web-based tool for accelerating discovery of gene regulatory networks using microarray co-expression data. BMC Bioinform. 10(1), 332 (2009)
Karolchik, D., et al.: The UCSC table browser data retrieval tool. Nucleic Acids Res. 32(suppl–1), D493–D496 (2004). https://doi.org/10.1093/nar/gkh103
Kel-Margoulis, O., Kel, A., Reuter, I., Deineko, I., Wingender, E.: TRANSCompel: a database on composite regulatory elements in eukaryotic genes. Nucleic Acids Res. 30, 332–334 (2002)
Kerren, A., Kucher, K., Li, Y.F., Schreiber, F.: Biovis explorer: a visual guide for biological data visualization techniques. PLOS ONE 12(11), 1–14 (2017). https://doi.org/10.1371/journal.pone.0187341
Kharumnuid, G., Roy, S.: Tools for in-silico reconstruction and visualization of gene regulatory networks (GRN). In: 2015 Second International Conference on Advances in Computing and Communication Engineering (ICACCE), pp. 421–426. IEEE (2015)
Kirlew, P.W.: Life science data repositories in the publications of scientists and librarians. Issues Sci. Technol. Libr. 65 (2011)
Krupp, M., Marquardt, J.U., Sahin, U., Galle, P.R., Castle, J., Teufel, A.: RNA-Seq Atlas - a reference database for gene expression profiling in normal tissue by next-generation sequencing. Bioinformatics 28(8), 1184–1185 (2012). https://doi.org/10.1093/bioinformatics/bts084
Lizio, M., et al.: Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol. 16(1), 22 (2015). https://doi.org/10.1186/s13059-014-0560-6
Longabaugh, W.J., Davidson, E.H., Bolouri, H.: Visualization, documentation, analysis, and communication of large-scale gene regulatory networks. Biochim. Biophys. Acta (BBA) - Gene Regul. Mech. 1789(4), 363–374 (2009). http://www.sciencedirect.com/science/article/pii/S1874939908001624
Margolin, A.A., et al.: ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinform. 7(1), S7 (2006)
Matharu, N., Ahituv, N.: Minor loops in major folds: enhancer-promoter looping, chromatin restructuring, and their association with transcriptional regulation and disease. PLOS Genet. 11(12), 1–14 (2015). https://doi.org/10.1371/journal.pgen.1005640
Meckbach, C., Tacke, R., Hua, X., Waack, S., Wingender, E., Gültas, M.: PC-TraFF: identification of potentially collaborating transcription factors using pointwise mutual information. BMC Bioinform. 16(1), 400 (2015). https://doi.org/10.1186/s12859-015-0827-2
Mora, A., Sandve, G.K., Gabrielsen, O.S., Eskeland, R.: In the loop: promoter-enhancer interactions and bioinformatics. Brief. Bioinform. 17(6), 980–995 (2016). https://doi.org/10.1093/bib/bbv097
O’Donoghue, S.I., et al.: Visualizing biological data - now and in the future. Nature Methods 7(3), S2 (2010)
Petryszak, R., et al.: Expression Atlas update - an integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res. 44(D1), D746–D752 (2015)
Ren, J., Lu, J., Wang, L., Chen, D.: Data visualization in bioinformatics. Adv. Inf. Sci. Serv. Sci. 4(22) (2012)
Roy, S., Bhattacharyya, D.K., Kalita, J.K.: Reconstruction of gene co-expression network from microarray data using local expression patterns. BMC Bioinform. 15(7), S10 (2014)
Schaffter, T., Marbach, D., Floreano, D.: Genenetweaver: in silico benchmark generation and performance profiling of network inference methods. Bioinformatics 27(16), 2263–2270 (2011)
Smoot, M.E., Ono, K., Ruscheinski, J., Wang, P.L., Ideker, T.: Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27(3), 431–432 (2010)
Sonawane, A.R., et al.: Understanding tissue-specific gene regulation. Cell Rep. 21(4), 1077–1088 (2017)
Steuernagel, L., Wiese, L., Gültas, M.: Repository visualization of dynamic biological networks. https://github.com/azifiDils/Visualization-of-DynamicBiological-Networks-
Tripathi, S., Dehmer, M., Emmert-Streib, F.: NetBioV: an R package for visualizing large network data in biology and medicine. Bioinformatics 30(19), 2834–2836 (2014)
Wang, M., et al.: LegumeGRN: a gene regulatory network prediction server for functional and comparative studies. PloS One 8(7), e67434 (2013)
Whitfield, T.W., et al.: Functional analysis of transcription factor binding sites in human promoters. Genome Biol. 13(9), R50 (2012). https://doi.org/10.1186/gb-2012-13-9-r50
Wiese, L.: Advanced Data Management for SQL, NoSQL. Cloud and Distributed Databases, DeGruyter/Oldenbourg (2015)
Wiese, L., Schmitt, A.O., Gültas, M.: Big data technologies for DNA sequencing. In: Sakr, S., Zomaya, A. (eds.) Encyclopedia of Big Data Technologies. Springer, Cham (2018). https://doi.org/10.1007/978-3-319-63962-8
Acknowledgements
Chimi Wangmo participated in the preparation of this article while visiting the University of Göttingen with a Go International Plus scholarship by the Erasmus+ Key Action of the European Commission.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Wiese, L., Wangmo, C., Steuernagel, L., Schmitt, A.O., Gültas, M. (2019). Construction and Visualization of Dynamic Biological Networks: Benchmarking the Neo4J Graph Database. In: Auer, S., Vidal, ME. (eds) Data Integration in the Life Sciences. DILS 2018. Lecture Notes in Computer Science(), vol 11371. Springer, Cham. https://doi.org/10.1007/978-3-030-06016-9_3
Download citation
DOI: https://doi.org/10.1007/978-3-030-06016-9_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-06015-2
Online ISBN: 978-3-030-06016-9
eBook Packages: Computer ScienceComputer Science (R0)