Abstract
Gene expression data of differentiating cells, galaxies distributed in space, and earthquake locations, all share a common property: they lie close to a graph-structured topology in their respective spaces [1, 4, 9, 10, 20], referred to as one-dimensional stratified spaces in mathematics. Often, the uncovering of such topologies offers great insight into these data sets. However, methods for dimensionality reduction are clearly inappropriate for this purpose, and also methods from the relatively new field of Topological Data Analysis (TDA) are inappropriate, due to noise sensitivity, computational complexity, or other limitations. In this paper we introduce a new method, termed Local TDA (LTDA), which resolves the issues of pre-existing methods by unveiling (global) graph-structured topologies in data by means of robust and computationally cheap local analyses. Our method rests on a simple graph-theoretic result that enables one to identify isolated, end-, edge- and multifurcation points in the topology underlying the data. It then uses this information to piece together a graph that is homeomorphic to the unknown one-dimensional stratified space underlying the point cloud data. We evaluate our method on a number of artificial and real-life data sets, demonstrating its superior effectiveness, robustness against noise, and scalability. Code related to this paper is available at: https://bitbucket.org/ghentdatascience/gltda-public.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Motivation. Identifying and visualizing graph-structured topologies underlying point cloud data sets is a non-trivial and active topic of research, with known applications in many fields of science, such as biology, physics, geology, geography, and computer science [1, 4, 10, 19, 20].
When the underlying graph-structured topology of D is well-modeled by a proximity graph, counting connected components in induced subgraphs suffices to learn topological structures locally, as well as the presence of cycles (see Algorithm 1 in Sect. 2, \(|D|=873\), \(\epsilon =3.5\), \(r=3\), comp. time: 0.43 s). By using these identified local topologies, we are able to reconstruct a graph homeomorphic to the underlying space (see Algorithm 2 in Sect. 3, \(r'=4\), comp. time: 8.04 s).
E.g., consider data of differentiating cells in a high-dimensional expression space. The way in which different cell stages are interconnected during cell differentiation can be represented by means of a graph (which may contain cycles) in the expression space, such that each of the differentiating cells lie close to it. More formally, the point cloud data approaches a topological structure homeomorphic to (i.e., obtainable from by ‘bending’ and ‘stretching’) the embedding of a corresponding graph in the expression space. In the mathematical literature, such an embedding is know as a one-dimensional stratified space (in this paper referred to as a graph-structured topology), composed of 0-D strata (here called the vertices) and 1-D linear strata (here called the edges or loops), glued together in a particular way.
A toy data set D is shown in Fig. 1 for illustration. Here the colored dots represent data points, and the black dots and lines represent vertices and edges of the graph-structured topology. The different colors express both local and global topological information, which we simply refer to as local topologies. E.g., near the center of the ‘8 component’, points are marked by a (4, 2) local topology, meaning four branches emerge from this location, and induce two cycles by convergence. We will formally explain this below. As this data set is 2-dimensional, its graph-structured topology is readily noticed. However, it is clear that such topologies, in high-dimensional data, are hard to uncover, and standard dimensionality reduction techniques will fail in all but the most trivial cases.
The emergent area of Topological Data Analysis (TDA) [5], which aims to understand the shape of data [23], seems to be the obvious approach to handle this problem. Its power for uncovering the underlying topology of data sets has been demonstrated in several recent works [3, 7, 13, 19,20,21]. However, TDA methods designed for this problem, such as Mapper [19, 20], local persistent (co)homology [11, 21], functional persistence [6], and metric graph reconstruction [1], are either computationally inefficient, restricted to specific graph-structured topologies, vulnerable to noise, or simply do not consider reconstructing the topology.
In this paper, we develop a novel method to fill this gap, under the name of Local Topological Data Analysis (LTDA). Investigating structures locally allows one to detect the degree, denoted \(\delta _0\), i.e., the number of branches emerging from a point, as well as the number of cycles, denoted \(\delta _1\), induced by the convergence of the same branches away from this point. LTDA provides methods for classifying data points according to their local topology \((\delta _0, \delta _1)\), identifying isolated, end-, edge- and multifurcation points, as well as cycles, by only tracking the number of connected components in graphs [2, 15] (Algorithm 1 in Sect. 2). Note that the discovery of cycles in such data using state-of-the-art TDA techniques requires the computation of the first order Betti number, the computation of which is challenging [24]. Combining the information retrieved by LTDA with clustering techniques allows for a fast reconstruction of the underlying graph-structured topology (Algorithm 2 in Sect. 3). These concepts are illustrated on Fig. 1.
Contributions
-
We develop a method, under the name of Local Topological Data Analysis (LTDA). This method allows us to detect isolated, end-, edge- and multifurcation points, as well as cycles, underlying data approaching graph-structured topologies, by merely counting the number of connected components in proximity graphs (Algorithm 1 in Subsect. 2.3).
-
We develop a framework that combines the information retrieved from LTDA with clustering techniques to reconstruct and visualize the unknown underlying topology of such data sets (Algorithm 2 in Sect. 3).
-
We clarify and empirically validate the usefulness of our methods on a variety of simulated and real data sets (Sects. 2, 3 and 4). We show that our methods are competitive with current state-of-the-art approaches in terms of results and computational efficiency.
-
We discuss how future research on the potential of LTDA may open up new possibilities to the set of TDA methods (Sect. 5).
2 LTDA of Graph-Structured Topologies
Given a Euclidean point cloud data set \(D\subseteq \mathbb {R}^n\) with an unknown underlying topological structure, we wish to investigate the global topology, i.e., the complete and unknown topological structure, by applying TDA to small patches of data, indicating (unknown) properties of the local topology. We start by showing how knowing both the local topological structures, as well as how these affect the global structure, may unravel graph-structured topologies. This leads to an algorithm proposed in this paper for identifying and locating multifurcation points and cycles in point cloud data approaching such topologies (Algorithm 1, Subsect. 2.3).
2.1 Overview: Illustrating the Idea Behind LTDA on a Toy Example
Here we first introduce LTDA in an intuitive and constructive way. We will do this by means of a simple two-dimensional toy data set. The used underlying topological structure of the toy data will show to be quite useful to understand the intuition behind Theorem 1 (Subsect. 2.2), which forms the foundation for the proposed approach of LTDA for graph-structured topologies (Subsect. 2.3).
A Toy Data Set. The toy data set \(D\subseteq \mathbb {R}^2\) we consider is the subset of the data illustrated in Fig. 1, that has the underlying topological structure of ‘the number 8’, illustrated in Fig. 2. Without going much into detail, an n-manifold is a topological spaceFootnote 1 locally resembling the Euclidean space of dimension n near every point on the space. There are essentially two (non-homeomorphic) connected 1-manifolds: the circle \(\mathcal {S}^1\) and the real line \(\mathbb {R}\). The underlying topology \(\tau \) of D is that of (homeomorphic to) two circles \(\mathcal {S}^1_1\) and \(\mathcal {S}^1_2\), intersecting in one singular point \(x\in \mathcal {S}^1_1\cap \mathcal {S}^1_2\).
The idea behind LTDA for data that approaches a graph-structured topology \(\tau =\mathcal {S}_1^1\cup \mathcal {S}_2^1\). For appropriate proximity graphs, one finds the underlying degree of a data point z (black) by counting the connected components in the graph induced by the intersection of a spherical shell and the data (green points), representing branches emerging from z. Convergence of these branches away from z indicates cycles through z, which may be identified by comparing the obtained degree with the number of connected components in the graph induced by the points away from z (blue and green points). (Color figure online)
The Idea Behind LTDA. One may assign a point \(y\in \tau \) to two classes: either \(y\ne x\) or \(y=x\). If \(y\ne x\), then y inherits its local topology from exactly one of the circles \(S^1_1\) or \(S^1_2\). As these are 1-manifolds, y has a neighborhood homeomorphic to \(\mathbb {R}\), or equivalently, to ]0, 1[. Removing any point c from ]0, 1[ breaks the interval into two disjoint connected components, as one can either move left or right from c in ]0, 1[. The same behavior occurs at y: starting from y, we can move into two directions, i.e., two branches emerge from y. If we would remove y from a neighborhood of y homeomorphic to ]0, 1[, then this neighborhood would break into two disjoint connected components as well. If \(y=x\), then four branches emerge from y, and removing y from a small neighborhood of y in \(\tau \) breaks the neighborhood into four components.
When a point cloud data set approaches a graph-structured topology, it reflects similar properties as that underlying topology. Consider the centered black data point \(z\in D\) in Fig. 2, representing the singular point x in the underlying topology \(\tau \). A neighborhood of x in \(\tau \) now corresponds to the points contained in a small open ball centered at z. Removing x from this neighborhood in \(\tau \) corresponds to removing points in an even smaller ball centered at z, strictly contained within the original ball. The points remaining in the spherical shell determined by these two concentric circles, or in general, hyperspheres, now represent the four components that result from removing x from a small neighborhood of x in \(\tau \) (green points in Fig. 2). Moreover, for an appropriate proximity graph constructed from D (see below and Fig. 2), the remaining points induce exactly four connected components in this graph. Hence, by only tracking the number of connected components in graphs [2, 15], we deduce the underlying degree \(\delta _0\), denoting the number of branches emerging from a data point.
Investigating the local topology of \(z\in D\) (black) by studying the underlying topology of \(B_{\mathbb {R}^2}(z,r)\cap D\) for increasing values of r. Points in \(B_{\mathbb {R}^2}(z,r)\cap D\) are marked in red (\(r = 1, 2, 3, 4\)), remaining points in blue. This method starts off well, but quickly becomes susceptible to the restrictions imposed by the underlying topology on paths. (Color figure online)
While classical approaches for TDA of data approaching graph-structured topologies stop at this point [1, 11], our concept of LTDA goes one step beyond. Not only are we interested in the local topology underlying a data point, i.e., the number of branches emerging from this point, but we are also interested in how this local topology affects the global topology. Consider again the singular point x in our discussed topology \(\tau \). As stated before, removing x from a small neighborhood of x breaks the neighborhood into four connected components, i.e., four branches emerge from x. However, moving further from x, two times two of these branches merge back together, and form cycles passing through x. As these branches merge back away from x, this implies that they must be connected in another way than through x. They are connected in the global topology even after removing x. Moreover, as removing x from a small neighborhood of x in \(\tau \) breaks the neighborhood into \(\delta _0=4\) components, but removing x from the full topological structure breaks the structure only into two connected components, the difference between these two denotes a practical lower bound on the number of convergences \(\delta _1 = \delta _0 - 2 = 2\) induced by the branches emerging from x (Theorem 1). In Fig. 2, this corresponds to subtracting the number of connected components induced by the points outside the smallest circle (green and blue points), from the number of connected components induced by the points in the spherical shell (green points). Hence, we may not only apply LTDA to identify the underlying local topology, i.e., the number of emerging branches, but we may as well identify cycles by studying how the local topology affects the global topology.
The Vietoris-Rips Complex. As D is a point cloud data set, it does not make much sense to talk exactly about the local topology of some point \(x\in D\) within the topological (normed vector) space \((D, \Vert \cdot \Vert )\), as this would be just a set of isolated points. However, for appropriate distance parameters \(\epsilon \in \mathbb {R}^+\), which may be found by means of persistent homology (Appendix A), the Vietoris-Rips complex
‘well-models’ topological behavior of the underlying topology \(\tau \) of D (Appendix A), and it makes more sense to talk about the local topology of a point \(\{x\}\in \mathcal {V}_{\epsilon }(D)\). The complex corresponds to the hypergraph induced by the cliques up to size \(\dim (D)+1\) of its graph ‘skeleton’, i.e., the graph consisting of all nodes from D and all edges \(\{v,w\}\in 2^D,\) where \(0<\Vert v-w\Vert <\epsilon \) (Fig. 2, \(\epsilon =3.5\)). We will also talk about the (Vietoris-Rips) graph \(\mathcal {V}_\epsilon (D)\) when referring to the skeleton of the complex, as we only consider simplicial 1-complexes, i.e., graphs in this paper.
Investigating the local topology of \(z\in D\) (black) by studying topological properties of \(\mathcal {V}_{0.3}(B_{\mathcal {V}_{0.3}(D)}(z, h+1))\) for increasing values of h. Vertices from \(\mathcal {V}_{0.3}(B_{\mathcal {V}_{0.3}(D)}(z, h))\) are marked in red, from \(\mathcal {V}_{0.3}(B_{\mathcal {V}_{0.3}(D)}(z, h+1)\backslash B_{\mathcal {V}_{0.3}(D)}(z, h))\) in green (\(h = 1, 10, 20, 27\)), and remaining points in blue. The underlying linear structure is preserved until all points are included at \(h=27\). (Color figure online)
A Metric for LTDA Derived from the Vietoris-Rips Graph. The open balls in Fig. 2 are drawn using the Euclidean metric, i.e., the balls denote sets
for some \(r>0\). Using this ‘original’ metric to investigate local topologies in \(\mathcal {V}_{\epsilon }(D)\) seems like a natural approach. However, in the general case, we may not be able to reach one point from another by following a straight line within the topological structure itself. In general, we are restricted to follow paths, corresponding to new distances defined by integrating over these when possible. Following this intuition, we ‘redefine’ the metric on D by defining the distance between two points as the distance within the graph \(\mathcal {V}_{\epsilon }(D)\). These geodesic distances, i.e., lengths of the shortest paths between nodes in the graph, are used to approximate the lengths of the shortest paths between the nodes’ projections on the underlying topology. This metric corresponds to new open balls in D, containing finitely many data points, and defined as
Figures 3 and 4 illustrate this for a point cloud data set approaching an ellipse.
Remark. We emphasize the difference between the (embedding of a) graph G underlying a point cloud data set D, and the Vietoris-Rips graph \(\mathcal {V}_{\epsilon }(D)\) constructed from D. These are generally non-homeomorphic in a graph-theoretical sense [16]. The unknown structure of G is often simple, with only a few multifurcation points and cycles. The known graph topology of \(\mathcal {V}_{\epsilon }(D)\) itself is often complex, with many multifurcation points and cycles present in the graph. This may be seen on the toy data set in Fig. 2. As a graph itself, \(\mathcal {V}_{\epsilon }(D)\) is quite complex, with many cycles and multifurcation points, i.e., nodes with degree at least equal to 3, whereas the underlying 8-structured topology of D is homeomorphic to the planar embedding of a graph with only two cycles and one multifurcation point. However, \(\mathcal {V}_{\epsilon }(D)\) is generally constructed such that it well-models particular topological behavior of G as discussed in Appendix A. Hence, Theorem 1 in Subsect. 2.2 will reside in the field of graph theory where we consider G, not \(\mathcal {V}_{\epsilon }(D)\). In Subsect. 2.3 the theorem will be translated into a data setting within the context of LTDA, i.e., for use on \(\mathcal {V}_{\epsilon }(D)\), by means of connected components.
2.2 Locally Analyzing a Graph Gives Global Insights
We now formalize the insights obtained from the discussion above in a graph-theoretical theorem. While this theorem applies to general graphs, in Subsect. 2.3, we show how it can be applied to proximity graphs representing the underlying topology of point cloud data. We assume graphs to be simpleFootnote 2, finite, and undirected, and that the reader is familiar with basic concepts of graph theory.
Notations. For a graph \(G=(V,E)\), we denote the number of connected components byFootnote 3 \(\beta _0(G)\), and the degree of a node \(v\in V\) by \(\delta _0(v)\). The degree of any edge \(e\in E\) is by definition \(\delta _0(e):=2\). If \(\alpha \in V\cup E\), we denote by \(G\backslash \alpha \) the graph that results from removing \(\alpha \) from G, as well as all edges incident to \(\alpha \) if \(\alpha \in V\).
Theorem for LTDA of Data Approaching Graph-Structured Topologies. The following theorem illustrates how the local topology of a node or an edge \(\alpha \) in a graph G, expressed by its degree \(\delta _0(\alpha )\), and how this local topology affects the connectedness of the global topology, expressed by the term \(\beta _0(G)-\beta _0(G\backslash \alpha )\), may be used to learn a practical lower bound on the number of cycles passing through \(\alpha \). Moreover, the theorem allows us to exactly determine whether a cycle passes through a node or an edge in a graph or not.
Theorem 1
Let \(G=(V,E)\) be a graph. Then for each \(\alpha \in V\cup E\), the number of cycles \(C\subseteq E\) passing through \(\alpha \) is bounded from below by
Moreover, for each \(\alpha \in V\cup E\), a cycle passes through \(\alpha \) iff \(\delta _1(\alpha )>0\).
Proof
The statements easily follows by induction from the well-known fact that inserting an edge into a graph either merges two connected components, or adds a cycle through that edge. Details are omitted for conciseness. \(\square \)
2.3 LTDA of Data Approaching Graph-Structured Topologies
To be applicable for LTDA of point cloud data approaching graph-structured topologies, we show how to translate Theorem 1 into a data setting. This will allow us to construct an algorithm identifying multifurcation points and cycles present in the underlying topology by merely counting the number of connected components in a proximity graph constructed from such data (Algorithm 1).
We again emphasize the difference between the (embedding of a) graph G underlying a point cloud data set D, and the simplicial complex \(\mathcal {V}_{\epsilon }(D)\) constructed from D. As remarked in Subsect. 2.1: these are generally non-homeomorphic in the graph-theoretical meaning. However, they approximate each other in terms of topological behavior as discussed in Appendix A.
Graph-Structured Topologies in a Data Setting. When a point cloud data set D approaches (the embedding of) a graph \(G=(V,E)\) in \(\mathbb {R}^n\) that is well-modeled by \(\mathcal {V}_{\epsilon }(D)\) for some \(\epsilon \in \mathbb {R}^+\), we may study the topology near \(x\in D\), represented by \(\alpha _x\in V\cup E\), by letting
-
\(\beta _0(G)\) correspond to \(\beta _0(\mathcal {V}_{\epsilon }(D))\),
-
\(\beta _0(G\backslash \alpha _x)\) correspond to \(\beta _0(\mathcal {V}_{\epsilon }(D\backslash B_{\mathcal {V}_{\epsilon }(D)}(x, r)))\),
-
\(\delta _0(\alpha _x)\) correspond to \(\beta _0(\mathcal {V}_{\epsilon }(B_{\mathcal {V}_{\epsilon }(D)}(x, r')\backslash B_{\mathcal {V}_{\epsilon }(D)}(x, r)))\),
for some \(0\le r<r'\) (see the discussion in Subsect. 2.1 and Fig. 2). All results in this paper were obtained by taking \(r'-1=r\in \{2,3\}\).
Hence, we may provide a mapping \(D\rightarrow \mathbb {N}\times \mathbb {N}:x\mapsto (\delta _0(x), \delta _1(x))\), expressing the underlying local topology at \(\alpha _x\), as well as a lower bound on the number of cycles through \(\alpha _x\), furthermore indicating whether or not a cycle passes through \(\alpha _x\) (Algorithm 1). We illustrate the use of this algorithm on an artificially constructed data set D based on the conference acronym, see Fig. 1.
E.g., the ‘ends’ of the four homeomorphic C, M, L and I-structured topologies are truthfully marked as (1,0) local topologies, i.e., structures resembling half-lines. The quotation mark is completely marked as having a (0,0) local topology, meaning this structure represents an isolated point. This shows that our algorithm may as well identify outlying points or areas, if the used proximity graphs models the underlying topology well. The (4,2) local topology in the 8-structured component marks an area with a local star-like topology with four legs, through which, in this case exactly, two cycles pass within the global topology.
Algorithm. For computational efficiency, the proposed algorithm marks neigbors of a node with a particular local topology with the same local topology. We implemented the algorithm such that nodes at a particular distance from another node are determined by a breadth-first search construction [2]. Hence, the total number of connected components in G is not needed to compute \(\delta _1\). If the inputted graph G has n vertices and m edges, where \(m=\mathcal {O}(\delta n)\) for some ‘average’ degree \(\delta \), the while loop will be executed \(\mathcal {O}(n/\delta )\) times. As each step in the loop can be executed in linear, i.e., \(\mathcal {O}(n+m)=\mathcal {O}(n+\delta n)\) time [2], the total complexity is \(\mathcal {O}(n^2)\).

Tuning \(\varvec{\epsilon }\) and \({\varvec{r}}\mathbf {.}\) The distance parameters \(\epsilon \) and r may usually be tuned by manual investigation. For all results in this paper, it was sufficient to investigate the use of either \(r= 2\) or \(r = 3\). Tuning \(\epsilon \) is more data dependent, and may be done by persistent homology as well (Figs. 13 and 14 in Appendix A). One may also integrate over different parameter ranges, which are bounded by the maximal pairwise distance for \(\epsilon \), and by the radius of the graph for r. Consequently, one inspects how well the reconstructed graph (Sect. 3) approximates the original graph, checking for a balance between reducing the Hausdorff distance, MSE, or metric distortion, (e.g., one may redefine distances as their projected distances on the reconstruction,) and reduction of the graph size, as also discussed in [1].
3 LTDA for Reconstructing Graph-Structured Topologies
In this section, we show the importance of LTDA for reconstructing the underlying topology. More concretely, we illustrate why the information retrieved by LTDA needs to be both stored and used, and why a simple ‘edge or no-edge’ classification as used in the metric graph reconstruction algorithm [1] may not always lead to optimal results for noisy samples. The latter method uses, similar to our approach, spherical shell clustering in a Vietoris-Rips graph to identify branching structures, but only classifies points according to \(\delta _0=2\) (edge) or \(\delta _0\ne 2\) (branch). The graph reconstruction is based on placing an edge between connected components of branch points, if they are both near one connected component of edge points. For further details on this method, we refer to [1].
Consider the simulated noisy two-dimensional data set D approaching a Y-structured topology with nonuniform density in Fig. 5. Our method of subgraph clustering (Algorithm 1) correctly infers the location of the (1,0) and (3,0) local topologies. However, due to the high amount of noise relative to the length of the branches, no (2,0) local topologies are detected. In this case, an ‘edge or no-edge’ classification as in [1] would lead to one connected component of branch points, of which the reconstructed graph [1] would be a single vertex.
Nevertheless, the (3,0) local topology ‘hints’ the presence of three surrounding branches. Simply clustering the (1,0) local topologies in their induced subgraph would not lead to three connected components, as two of the branches would not be separated (cluster 2 & 3 in Fig. 5). This is a straightforward consequence of the underlying topology: even when we remove the bifurcation point, the branches are still at distance 0 from each other. Inseparability of the branches may even occur for less noisy data with uniform density, when the distance parameter \(\epsilon \) was tuned too high. However, in this particular example there does not even exist a single distance value \(\epsilon \) for which the three clusters would be pairwise separable in their induced subgraph of \(\mathcal {V}_{\epsilon }(D)\), due to the nonuniform density.
Algorithm. A different clustering algorithm exploiting the information of the (3,0) local topology is needed. Applying hierarchical clustering (we use complete-linkage clustering unless stated otherwise), allows us to separate the points neighboring the (3,0) local topologies in three clusters (Fig. 5), leading to Algorithm 2 for reconstructing general underlying graph-structured topology. The pseudocode assumes the used graph G and distance object d stored in the output of Algorithm 1.

The pseudocode of Algorithm 2 allows for many variants in its implementation. E.g., many steps implicitly assume most pairwise distances defined by d to be unique, and we use the original Euclidean metric used to construct our proximity graph for Algorithm 1. We define the center of a set \(X\subseteq D\) as the data point \(c_X := \arg \min _{x\in X}(\max _{y\in Y} d(x,y))\), which leads to better results than the point closest to the mean in the case of nonuniform density. Representing the center in our current way works well for short patches of the underlying topology, but is less efficient for patches representing long and curvy trajectories (red graph in Fig. 6). Using a new metric defined by distances in the weighted graph \(\mathcal {V}_{\epsilon }(D)\), with the Euclidean lengths of the edges as weights, may lead to even better results for computing centers of long and curvy patches and (hierarchical) clustering into a given number of clusters, at the cost of computational efficiency. An alternative method is to use a breadth-first traversal to decompose long clusters representing edges into short and consecutive patches (black graph in Fig. 6, note that both graphs are nevertheless homeomorphic), or one may connect different centers by shortest paths as well. Isolated circles are separated into four components by starting a breadth-first traversal at a random point, dividing points according to low, medium, or high distance from the root, and dividing the points at medium distance into two separate components. Finally, we replace the representative point of a (1,0) component such that it is furthest from its adjacent center.
Tuning \(\varvec{\tilde{r.}}\) The distance parameter \(\tilde{r}\) may be either tuned manually (all results in this paper were obtained by using either \(\tilde{r}=r\) or \(\tilde{r}=r+1\), r being the distance parameter used to obtain the output of Algorithm 1), or tuned in an integration scheme as discussed in Subsect. 2.3. However, a new distance parameter \(\tilde{r}\) is not needed for components resembling isolated points, edges, cycles or multifurcating trees. This last observations follows from
for a tree \(T=(V, E)\) with \(|E|\ge 1\) and no vertices of degree 2 (these are irrelevant for representing the underlying topology). This implies that the union of points having either (1,0) or (2,0) local topologies must be clustered into \(|E|=\sum _{\begin{array}{c} v\in V\\ \delta _0(v)\ge 3 \end{array}}\delta _0(v)-|\{v\in V:\delta _0(v)\ge 3\}|+1\) components, where this number is computed with respect to the connected components with \(\delta _0\ge 3\). If the tree has at least one multifurcation point, all such obtained clusters of edges will be incident to at least one multifurcation point and represented by at least two nodes in the reconstructed graph topology. This allows for another variant of Algorithm 2 for tree-structured topologies: cluster the union of (1,0) and (2,0) local topologies in the obtained number of clusters, and connect each component with \(\delta _0\ge 3\) to all adjacent clusters of edges.
4 Experimental Results
Our method is validated on two more real point cloud data sets approaching graph-structured topologies. All our results were obtained using non-optimized R code on a basic laptop.
Reconstructed graphs of the earthquake data set by the method discussed in [1].
Earthquake Data. We considered a geological data set D of 1479 strong to great earthquakes (Richter magnitude \(M_L > 6.5\)), scattered across the world in the rectangular domain \([140,315]\times [-75,65]\) of (longitude, latitude)-coordinates (180\(^\circ \) were added to negative longitudes to obtain a continuous structure). The raw data is freely accessible from USGS Earthquake Search. A distance to measure [8] from the R-package TDA was used to remove most outliers (\(m0=0.1\)), keeping 1440 observations with DTM < 30. The local topologies were classified in 0.90 s (\(\epsilon = 10, r =2\)), after which the underlying graph was reconstructed in 4.16 s (\(\tilde{r}=r=2\)). Two clusters representing long edges were decomposed into respectively 15 and 5 consecutive patches, resulting in the graph depicted in black in Fig. 7, approximating the underlying graph-structured topology well.
We compared our method with the original underlying graph reconstruction method as discussed in [1], where parameters were tuned to capture the single self-loop present in the underlying topology. We used both the original Euclidean metric (Fig. 8, bottom left, 4 min 11 s), as well as the metric induced by the weighted graph \(\mathcal {V}_{10}(D)\) (Fig. 8, top right, 2 min 41 s), but were unable to retrieve the full underlying topology with either of the metrics.
Cell Trajectory Data. We considered a normalized expression data set D of 4647 manually analyzed bone marrow cells containing measurements of five surface markers (CD34, CD1632, CD117, CD127 & Sca1). These cells are known to differentiate from long-term hematopoietic stem cells (LT-HSC) into short-term hematopoietic stem cells (ST-HSC), which can in turn differentiate into either common myeloid progenitor cells (CMP) or common lymphoid progenitor cells (CLP) [10]. I.e., the topology underlying this data set is that of an embedding in \(\mathbb {R}^5\) of the graph depicted in Fig. 9. No data preprocessing was applied, and the Euclidean distance was used as the original metric. A PCA plot of the data is shown in Fig. 10. Comparing Figs. 9 and 10, we indeed note the presence of the Y-structured topology. However, it is clear that identifying this topology would be a crucial problem in absence of the cell labeling. Hence, our method may serve as a first step in the context of cell trajectory inference [4, 10], identifying the branching structure and different stages within a cell differentiation process. Our method classified local topologies in 15.55 s (\(\epsilon = r = 2\)), and used these to reconstruct the underlying topology in 5.46 s. Note that the local topology classes ((1,0) and (3,0)) imply an underlying tree-structured topology, and no new distance parameter \(\tilde{r}\) is needed for the graph-reconstruction. We inferred the exact same graph using both complete and McQuitty’s linkage. However, the labeling induced by using the latter method, of which the result is shown in Fig. 11, correlated slightly better with the original cell types. The obtained branch-assignments correlate well with the original assignments, except for, most notably, non-CLP cells near the base of the ST-HSC\(\rightarrow \)CLP branch assigned to the branch itself.
We again compared our method to the original method [1] using two metrics (Euclidean: 1 h 17 min, and induced by the weighted graph \(\mathcal {V}_{2}(D)\): 1 h 35 min), but were unable to capture the underlying topology, as these methods resulted in an isolated cycle in both cases (\(>98\%\) of the data was marked as branch point, remaining edge points were inseparable). We also compared our method with MapperFootnote 4 [19, 20], using the freely accessible tool from the R package TDAmapper. Experimenting with different filter functions, only the projection onto the first principal component allowed us to correctly infer the underlying topology in 11.85 s. However, this was a matter of luck, as the assignments induced by the Mapper graph correlate badly with the original assignments (Fig. 12).
5 Conclusion and Further Work
Applying clustering techniques to study local topologies, and how these affect the global topology, introduces new possibilities for learning graph-structured topologies underlying point cloud data sets, as one may even detect cycles without the need of 1-dimensional homology. Current state-of-the-art approaches for investigating local topological structures either do not bother with reconstruction techniques, are vulnerable to noise, or miss out on the fact that knowledge of the local topologies is crucial for reconstructing underlying graph-structured topologies. We combined both LTDA and reconstruction techniques in a simple and intuitive way, leading to a framework for reconstructing the underlying graph in many practical examples, improving both on the computational level as well as the obtained results compared to current state-of-the-art approaches.
Contrary to [1], we prioritized explaining and validating our method by means of empirical results on simulated and real data sets, rather than providing theoretical results guaranteeing the correctness of the reconstructed graph topology. Real data will most often violate the stated assumptions, and the ‘one-for-all’ parameter approach posed by these may not be suitable when extending our method to even more complex and high-dimensional data sets approaching graph-structured topologies with nonuniform noise. For this, one needs local parameter integration schemes, combining results from the the fields of TDA (e.g., persistent local homology [11]), statistics, and machine learning. This provides new research both on the mathematical and experimental level.
Persistent homology of a point cloud data set approaching an ellipse. (Top) Each bar represents a connected component in \(\mathcal {V}_{\epsilon }(D)\) for varying \(\epsilon \). The long persisting bar indicates that there is one connected component present in the underlying topological structure. (Bottom) Each bar represents one of the non-equivalent cycles in \(\mathcal {V}_{\epsilon }(D)\) for varying \(\epsilon \). The long persisting bar indicates that there is one cycle present in the underlying topological structure.
The resulting graph (skeleton of) \(\mathcal {V}_{\epsilon }(D)\) for one of the distance parameters \(\epsilon =0.3\) occurring at both persisting bars in Fig. 13 (edges in black). The uniform (2,1) local topology indicates a cycle (see Subsect. 2.3, \(r=2\), comp. time: 0.14 s), and allows us to reconstruct the underlying topology (edges in red, see Sect. 3, comp. time: 0.17 s). (Color figure online)
Notes
- 1.
Formally, a topological space is an ordered pair \((X,\tau )\), where X is a set and \(\tau \) is a collection of subsets of X, satisfying particular axioms. The elements of \(\tau \) are called open sets and the collection \(\tau \) is called a topology on X. In this paper, we abuse notation for simplicity, and use \(\tau \) to refer to the set \(X=\bigcup \tau \).
- 2.
Loops and parallel edges may be subdivided without changing the graph topology.
- 3.
We maintain the terminology of homology, where \(\beta _0\) refers to the zeroth Betti number.
- 4.
Mapper uses a filter \(f:D\rightarrow \mathbb {R}^d\) that maps the data to a lower-dimensional space \(\mathbb {R}^d\) (usually \(d\in \{1,2\}\)), builds a grid of overlapping bins (intervals for \(d=1\), squares for \(d=2\), ...) on top of \(\mathbb {R}^d\), clusters the preimage \(f^{-1}(B)\) for each bin B, and connects clusters based on the overlap of the data and bins. This method results in the construction of a graph meant to resemble the unknown underlying topology, and has shown it may reveal a Y-structured topology in expression data before. For such an example and further details on the Mapper algorithm, we refer to [19].
References
Aanjaneya, M., Chazal, F., Chen, D., GLisse, M., Guibas, L., Morozov, D.: Metric graph reconstruction from noisy data. Int. J. Comput. Geom. Appl. 22(04), 305–325 (2012)
Bernhard, K., Vygen, J.: Combinatorial Optimization: Theory and Algorithms. Springer, Heidelberg (2012). https://doi.org/10.1007/3-540-29297-7
Cámara, P.G., Rosenbloom, D.I.S., Emmett, K.J., Levine, A.J., Rabadán, R.: Topological data analysis generates high-resolution, genome-wide maps of human recombination. Cell Syst. 3(1), 83–94 (2016)
Cannoodt, R., Saelens, W., Saeys, Y.: Computational methods for trajectory inference from single-cell transcriptomics. Eur. J. Immunol. 46(11), 2496–2506 (2016)
Carlsson, G.: Topology and data. Bull. Am. Math. Soc. 46(2), 255–308 (2009). https://doi.org/10.1090/S0273-0979-09-01249-X
Carlsson, G.: Topological pattern recognition for point cloud data (2013)
Carlsson, G., Ishkhanov, T., de Silva, V., Zomorodian, A.: On the local behavior of spaces of natural images. Int. J. Comput. Vis. 76(1), 1–12 (2008)
Chazal, F., Cohen-Steiner, D., Mérigot, Q.: Geometric inference for measures based on distance functions (2009)
Choi, E., Bond, N.A., Strauss, M.A., Coil, A.L., Davis, M., Willmer, C.N.A.: Tracing the filamentary structure of the galaxy distribution at z \(\sim \) 0.8. Mon. Not. R. Astron. Soc. 406(1), 320–328 (2010)
De Baets, L., Van Gassen, S., Dhaene, T., Saeys, Y.: Unsupervised trajectory inference using graph mining. In: Angelini, C., Rancoita, P.M.V., Rovetta, S. (eds.) CIBB 2015. LNCS, vol. 9874, pp. 84–97. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-44332-4_7
Fasy, B.T., Wang, B.: Exploring persistent local homology in topological data analysis. In: 2016 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), pp. 6430–6434 (2016)
Ghrist, R.: Barcodes: the persistent topology of data. Bull. (New Ser.) Am. Math. Soc. 45(107), 61–75 (2008)
Giusti, C., Ghrist, R., Bassett, D.S.: Two’s company, three (or more) is a simplex. J. Comput. Neurosci. 41(1), 1–14 (2016)
Hatcher, A.: Algebraic Topology. Cambridge University Press, Cambridge (2002)
Hopcroft, J.E., Ullman, J.D.: Set merging algorithms. SIAM J. Comput. 2(4), 294–303 (1973)
Lapaugh, A.S., Rivest, R.L.: The subgraph homeomorphism problem. J. Comput. Syst. Sci. 20(2), 133–149 (1980)
Medina, P., Doerge, R.: Statistical methods in topological data analysis for complex, high-dimensional data. In: Annual Conference on Applied Statistics in Agriculture (2015)
Nanda, V., Sazdanović, R.: Simplicial models and topological inference in biological systems. In: Jonoska, N., Saito, M. (eds.) Discrete and Topological Models in Molecular Biology. NCS, pp. 109–141. Springer, Heidelberg (2014). https://doi.org/10.1007/978-3-642-40193-0_6
Nicolau, M., Levine, A.J., Carlsson, G.: Topology based data analysis identifies a subgroup of breast cancers with a unique mutational profile and excellent survival. Proc. Nat. Acad. Sci. 108(17), 7265–7270 (2011)
Rizvi, A.H., et al.: Single-cell topological RNA-seq analysis reveals insights into cellular differentiation and development. Nat. Biotechnol. 35, 551–560 (2017)
Wang, B., Summa, B., Pascucci, V., Vejdemo-Johansson, M.: Branching and circular features in high dimensional data. IEEE Trans. Visual. Comput. Graph. 17, 1902–1911 (2011)
Wang, K.: The basic theory of persistent homology (2012)
Wasserman, L.: Topological data analysis. Ann. Rev. Stat. Appl. 5(1), 501–532 (2018)
Zomorodian, A., Carlsson, G.: Computing persistent homology. Discrete Comput. Geom. 33(2), 249–274 (2005)
Acknowledgments
This work was funded by the ERC under the European Union’s Seventh Framework Programme (FP7/2007-2013) / ERC Grant Agreement no. 615517, and the FWO (G091017N, G0F9816N).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
A Background on TDA: Persistent Homology
A Background on TDA: Persistent Homology
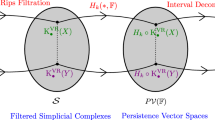
Finding an appropriate proximity graph is a crucial step for our method, as it identifies the number of emerging branches by counting the connected components in subgraphs induced by the intersection of our data and spherical shells (Fig. 2). Our choice of using Vietoris-Rips graphs is not arbitrary, as experimental results have shown that these are far more useful for our method than a wide variety of other proximity graphs, such as, e.g., k-nearest neighbor graphs. Moreover, Rips-graphs are well studied within the field of TDA, more concretely persistent homology, allowing to appropriately tune the distance parameter \(\epsilon \).
Persistent Homology [12, 22] tracks the (dis)appearance of distinct shape features across a filtration, i.e., a sequence of simplicial complexes [14]
constructed from a point cloud data set D embedded in a metric space, for an increasing sequence of parameters \(\epsilon _1,\ldots ,\epsilon _n\). By evaluating how long certain features exist, one is able to deduce topological invariants of its underlying topological structure [17]. The evolution of these (dis)appearing shape features may be modelled by means of barcodes, computed by methods of linear algebra [18, 24]. The number of bars occuring at a fixed value of \(\epsilon \) denotes the k-th Betti number, i.e., the number of k-dimensional holes, at the point \(\epsilon \) in the filtration. Long bars resemble topological features that ‘persist’ for many consecutive values \(\epsilon _i,\epsilon _{i+1},\ldots ,\epsilon _j\), and indicate features of the underlying topology of point cloud data. See Fig. 13, where the used filtration consists of Vietoris-Rips complexes.
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Vandaele, R., De Bie, T., Saeys, Y. (2019). Local Topological Data Analysis to Uncover the Global Structure of Data Approaching Graph-Structured Topologies. In: Berlingerio, M., Bonchi, F., Gärtner, T., Hurley, N., Ifrim, G. (eds) Machine Learning and Knowledge Discovery in Databases. ECML PKDD 2018. Lecture Notes in Computer Science(), vol 11052. Springer, Cham. https://doi.org/10.1007/978-3-030-10928-8_2
Download citation
DOI: https://doi.org/10.1007/978-3-030-10928-8_2
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-10927-1
Online ISBN: 978-3-030-10928-8
eBook Packages: Computer ScienceComputer Science (R0)