Abstract
We present a method for automatically determining the position and orientation of the articular marginal plane (AMP) of the proximal humerus in computed tomography (CT) images without segmentation or hand-crafted features. The process is broken down into 3 stages. Stage 1 determines a coarse estimation of the AMP center by sampling patches over the entire image and combining predictions with a novel kernel density estimation method. Stage 2 utilizes the estimate from stage 1 to focus on a smaller sampling region and operates at a higher images resolution to obtain a refined prediction of the AMP center. Stage 3 focuses patch sampling on the region around the center obtained at stage 2 and regresses the tip of a vector normal to the AMP which yields the orientation of the plane. The system was trained and evaluated on 27 upper arm CTs. In a 4-fold cross-validation the mean error in estimating the AMP center was \(1.30\,{\pm }\,0.65\) mm and the angular error for estimating the normal vector was \(4.68\,{\pm }\,2.84^\circ \).
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Shoulder arthroplasty is a common orthopaedic procedure indicated in certain cases of primary and secondary degenerative conditions. The annual rate of shoulder arthroplasty has been increasing with a steeper increase in total shoulder arthroplasty (TSA) compared to hemiarthroplasty since the early 2000s. In 2008, an estimated 46,951 procedures were performed in the USA (20,178 hemiarthroplasties and 26,773 total shoulder arthroplasties) [1]. Both of these procedures require replacing the humeral joint surface with TSA addressing the glenoid surface as well. This paper focuses on determining the location and orientation of the humeral head.
Preservation of articular anatomy with the purpose of maintaining physiologic soft tissue tension is the motivation behind humeral implant design. Traditional long-stemmed monobloc humeral implants have, for the most part, been replaced by modular versions and more recently short-stemmed, stemless, or resurfacing implants have gained popularity [2]. These modern implants allow adjustments that can fit the implant to match the anatomy encountered intraoperatively. They typically rely on resection of the humeral head along the anatomic neck, which is approximated geometrically by the articular marginal plane (AMP). In many cases this resection is done freehand intraoperatively; however, recognition of the importance of accurately reconstructing the humeral head anatomically is becoming more relevant, particular with short-stemmed, stemless, and resurfacing implants [2,3,4].
Key dimensions of the proximal humerus: GT = greater tuberosity, HA = humeral axis, HI = humeral head inclination, HV = humeral head version, LE = lateral epicondyle, LT = lesser tuberosity, ME = medial epicondyle. Figure (c) shows the hand-selected points defining the articular marginal plane in blue as well as the calculated ground truth center and normal in green. (Color figure online)
A high variability exists in the anatomy of the proximal humerus, typical accepted mean values summarized by Keener et al. [2] are shown in Fig. 1 and Table 1. Our focus will be defining where the center of the AMP is and its orientation in space. Further work would be required to define the humeral axis and epicondylar axis, from which the inclination and version could be defined. This provides motivation for today’s modular systems which can be adapted to a wide range of anatomical variations.
With modular systems, often a detailed preoperative analysis of the anatomic dimensions is not performed except in cases of extreme deformity or small size where special order or patient-specific components may be required. Simple templating using X-rays and scaled two-dimensional (2D) drawings of implants to visually confirm restoration of anatomy is often all that is performed preoperatively [3]. The surgeon can adjust the exact orientation and size components intraoperatively and determine appropriate fit by soft tissue assessment. The importance of the exact level of accuracy of the humeral component parameters is not well defined; however, it is suggested that restoration of the physiologic anatomy and forces would provide most success in restoring the kinematics of the shoulder and reducing shear stresses on the glenoid component [2, 5]. Accurate measurement of the AMP will likely become more important in cases such as resurfacing and stemless implants where the implant positioning is based directly off of the AMP.
Most implant companies provide surgeons with planning software that allows overlay of implant models on three-dimensional (3D) computed tomography (CT) data. Manual positioning of the virtual components can allow the surgeon to determine the appropriate size and position of components preoperatively, as well as to determine reaming and cutting trajectories. The information from these systems can currently also be used to produce patient specific guides that improve reproducibility of glenoid instrumentation guide pins [3].
Previous work has defined the AMP by manually selecting points on the anatomic neck on CT data and producing a best-fit plane to this [6, 7]. This is time consuming and its accuracy is prone to inter-observer variations [8]. Recently, Tschannen et al. [9] sought to automate the process using a random forest-based method. They compared their method to a manually-assisted atlas-based method and were able to improve accuracy.
Automated regression of landmarks using deep learning methods has seen recent success in several applications [10,11,12]. Automatic and computer-assisted techniques for determination of the glenoid parameters have been investigated and shown success in providing accurate information for preoperative planning [13,14,15]. The humeral head remains relatively neglected, with Tschannen et al. [9] being the only study identified regarding automatic parameter recovery. An attempt to create a system to improve the accuracy of automatic determination of the AMP utilizing fully convolutional neural networks will be explored. Our method will rely on a CT scan cropped roughly such that it must include the humeral head and from this predict the location and orientation of the AMP automatically.
2 Materials and Methods
2.1 Method Overview
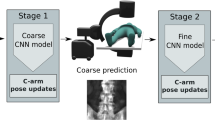
The aim of the project is to develop a deep learning-based method to automatically determine the AMP given an upper arm CT scan without segmentation or hand-crafted features. To fully define the AMP we require a point in the plane and a vector normal to it giving the orientation. We propose a 3-stage, multiscale, cascaded system to achieve this. Each stage samples patches from the image and predicts an offset from the patch location to the desired landmark. The results are combined to form a prediction for the landmark at each stage. The first stage processes patches from the CT volume at a low resolution and combines them to predict a rough estimate for the center of the AMP. The second stage refines the center estimate by running at a higher resolution and focusing training at a region of interest (ROI) centered at the stage 1 prediction. Finally, the third stage runs at the higher resolution to predict the tip of the normal vector thus giving all the information required to define the AMP.
2.2 Data Description
We used 27 cropped CT scans of right shoulders from the previous work of Tschannen et al. [9]. The data were collected from the Institutes for Forensic Medicine of the Universities of Bern and Zurich, Switzerland. The CT scanners used were a Siemens Emotion 6® and a Siemens Somaton Definition Flash®. The cropped images typically had a field of view \(228.6\,{\times }\,228.6\,{\times }\,450\) mm (covering the area used clinically for assessing the upper arm) and a typical resolution of \(1.27\,{\times }\,1.27\,{\times }\,0.6\) mm though there were some scans that varied slightly from these parameters. All images were resampled into a isometric low and high resolution voxel size of \(1.25\,{\times }\,1.25\,{\times }\,1.25\) mm and \(0.6\,{\times }\,0.6\,{\times }\,0.6\) mm respectively.
The AMP was defined by 12 manually picked pointsFootnote 1 along the margin where articular cartilage transitions to bone. From these points our regression targets, the AMP center and the tip of the normal vector, were derived. The original 12 points were shifted to have a mean at the origin. Using singular value decomposition we obtained the orientation of this plane, yielding the normal. Performing least-squares fitting to a circle of the points projected into this plane defines the AMP center. The center and normal vector were then shifted back to the original location using the original mean.To define a single point for the patch-based regression, the tip of the normal vector was defined as the point where a normal emanating from the center of the AMP intersected the surface of the humeral head. The ground truth points overlaid on a 3D view of the proximal humerus are shown in Fig. 1(c).
2.3 Network Architecture
Inspired by the landmark regression FCN introduced in our previous work [10], here we designed patch-based FCNs to solve our problem. More specifically, we opted to utilize a multiscale approach with the regression split into three stages, each stage processes multiple patches in prediction mode and uses a modified kernel density estimation (KDE) to combine the information into a single prediction. The architecture defining our system is shown in Fig. 2.
The network used is shown with the output being the offset from the patch corner to the center of the articular marginal plane. Layer dimensions given in the form: channels @ patch shape. Stages 2 and 3 use the same layout but add an additional repeat of the convolution/convolution/maxpool structure to match the dimensions.
Stage 1. This low resolution stage utilizes an input patch size of \(32\,{\times }\,32\,{\times }\,32\) voxels at a voxel size of \(1.25\,{\times }\,1.25\,{\times }\,1.25\) mm. It generates a rough estimate of the AMP center location, which allows for a refinement at stage 2. It begins with a scheme repeated three times consisting of two 3D convolutions (each with a kernel of \(3\,{\times }\,3\,{\times }\,3\), a stride of 1, batch normalization, and a rectified linear unit, ReLU, activation) followed by max pooling (with size \(2\,{\times }\,2 \,{\times }\,2\) and a stride of 2). Next a convolution with kernel \(4\,{\times }\,4\,{\times }\,4\), stride 1, batch normalization, and an hyperbolic tangent (tanh) as activationFootnote 2 reduces the patch dimensions to \(1\,{\times }\,1\,{\times }\,1\). Another convolution with kernel \(1\,{\times }\,1\,{\times }\,1\), stride 1, batch normalization, and a tanh activation reduces the patch dimensions to \(1\,{\times }\,1\,{\times }\,1\). Finally, a convolution with kernel \(1\,{\times }\,1\,{\times }\,1\), stride 1, no batch normalization, and no activationFootnote 3 reduces the patch dimensions to 3 values representing the three coordinates of the displacement from the patch location to the target landmark.
Stages 2 and 3. These higher resolution stages utilize the same network structure as stage 1 with the exception that they take as input a patch size of \(64\,{\times }\,64\,{\times }\,64\) voxels at a voxel size of \(0.6\,{\times }\,0.6\,{\times }\,0.6\) mm in order to produce a regression with higher accuracy. To divide the larger patch size to the same output there are 4 repetitions of the input stages instead of 3. Stage 3 utilizes the same network definition as stage 2; however, it is trained to regress the tip of the normal vector instead of the center of the AMP.
2.4 Training
Previous work has suggested that limiting patch selection to points on edges has the potential to improve training time and accuracy [10, 16]. We adopted this by generating Canny edge maps for each image, sampling patches only from the voxels located on edges. Additionally, the region nearer to the humeral head is likely to have more relevant information on its pose [9], so we designed a new sampling strategy to sample more points in regions nearer to the humeral head as described below.
During training an equal number of patches are sampled from each spherical shell centered at the ground truth center with a smaller region of interest in each subsequent stage. During testing stage 1 samples randomly from the entire volume and generates a rough center prediction, \(c_1\). Stage 2 prediction uses shells centered at \(c_1\) to predict \(c_2\) which is used in turn as the center for the sampling shells in stage 3.
During training all patches were obtained from a spherical region of radius \(r_{\max }\) separated into shells of equal width, \(r_{\text {shell}}\), centered at the ground truth center of the AMP as illustrated in Fig. 3. Each batch consisted of a number of samples, \(n_s\), from a single image divided equally among the shells so that more patches were sampled from regions nearer to the center. The parameters for sampling for each stage are listed in Table 2.
During each epoch, each image was visited once in a newly randomized order and a different random sampling of patches was obtained. The mean-squared error loss function was used representing the Euclidean distance between predicted displacement and the ground truth displacement. The Adam optimizer algorithm with an exponential decay of the learning rate was employed [17]. Each stage is trained independently.
2.5 Testing
Testing proceeds in a cascaded fashion as shown in Fig. 4. Given an unseeen CT volume, stage 1 samples patches uniformly over the entire volume and generates a prediction at the lower resolution for the center of the AMP. This prediction is used as a center for the spherical sampling ROI in stage 2, concentrating the higher resolution patches at the region around the humeral head. From stage 2 we obtain a more accurate estimate of the AMP center which we also use as the ROI center for stage 3 sampling. Stage 3 finally produces a prediction for the normal vector tip. Sampling is illustrated in Fig. 3 and the parameters used are in Table 2.
At each stage 1024 patches are sampled and processed to generate a single prediction. The network returns a 3D offset vector to the predicted landmark location for each processed patch. Each patch in the sample set has a known location and thus generates an independent prediction for the location of the landmark. To improve the accuracy, the independent predictions are combined using an approximate KDE to generate a 3D probability map as described below.
Fast KDE Implementation. Typical KDE implementations are computationally intensive, a novel algorithm was implemented to generate a probability map by only calculating the kernel to 2 standard deviations along each direction. The standard deviation for each direction was approximated from the covariance in the distribution of landmark locations from the prediction sample set. This clipped Gaussian kernel was added to a 3D array of zeros the same size as the image centered at each prediction location generating a non-normalized approximate probability distribution. The location of the maximum value in this array was taken as the prediction of the landmark. The typical appearance of a prediction at each stage is shown in Fig. 5.
2.6 Implementation Details
The network described was trained and tested using Tensorflow 1.5 [18] in Python 3.6.5 on a Tesla 1080 Ti GPU using an Ubuntu Linux 16.04 workstation with an Intel Core i7-7700 CPU at 3.60 GHz and 32 GB RAM.
2.7 Experimental Design
We evaluated the accuracy of the present approach using a standard 4-fold cross-validation experiment. To this end, the set of 27 images provided was split into 3 groups of 7 images and 1 group of 6. For every fold of study, 3 out of 4 groups of data were used for training and the left-out group were used for testing. Stage 1 and 2 were trained 500 epochs and stage 3 was trained 100 epochs. The accuracy was evaluated by comparing the prediction for each of the images as described in Sect. 2.5 to the corresponding ground truth.
The error in the center of the AMP and vector tip predictions are defined as an L2 distance from the prediction to the associate ground truth. The angular error is determined by solving the cosine relationship for the angle between the predicted normal and the ground truth \(\theta \):
3 Results
The mean error for estimating the center of the AMP is \(1.30\,{\pm }\,0.65\) mm. The mean angular error was \(4.68\,{\pm }\,2.84^\circ \). A scatter plot showing the distribution for each prediction grouped by fold is shown in Fig. 6. Figures 6 and 7 demonstrate the distribution of our error measurements for the center of the AMP, the vector tip, and the angular error, respectively. In order to compare the estimation uncertainty of different quantities, we calculated the coefficient of variations (CV) for each quantity. We found \(CV_{center}\,{=}\,50\%\), \(CV_{vector}\,{=}\,42.1\%\), and \(CV_{angular}\,{=}\,60.7\%\), respectively, suggesting higher uncertainty in our angular error results. The uncertainty in estimating the normal vector is increased by the fact that we are compounding the error of the location of both the AMP center and the normal tip (calculated by 2 separate networks) when we compute the normal vector (Table 3).
4 Discussion
Accurate location and orientation of the AMP are key to planning the resection of the humeral head in both TSA and hemiarthroplasty. The level of accuracy needed in final humeral head orientation for a successful outcome is not fully defined; however, it is certainly an important factor in preserving the anatomical orientation of the proximal humerus which is key to successful kinematics and glenoid loading [2, 5]. Modern modular humeral implants as well as short-stemmed, stemless, and resurfacing techniques are focus attention on anatomical replacement of the humeral head [2]. To date, more work has been directed toward computer-assisted planning methods for the glenoid component. The only automated method for determining the parameters of AMP identified was Tschannen et al. [9].
Our results of a mean error for the center of the AMP of \(1.30\,{\pm }\,0.65\) mm and a mean angular error of \(4.68\,{\pm }\,2.84^\circ \) are an improvement on the prior work. Additionally, our choice to restrict the ROI to an area around the humeral head in training seems to have supported Tschannen et al. [9] in assuming this is the area containing the most relevant information for determining the parameters of the AMP.
The data from this technique also yields the height of the humeral head directly as the length of the normal vector, though this was not assessed for accuracy at this time. Additional information, such as the radius of the AMP, could be regressed with the given ground truth information simply by regressing an additional point in a similar fashion to stage 3. With this information one could fully define the parameters of humeral head. An additional system could be developed to define the orientation of the humeral shaft, combined with our approach this could fully define the humeral implant parameters as per Fig. 1. Our work could be integrated into computer-assisted surgery systems to provide a cutting plan for resection of the AMP. Deep learning methods offer an extendible, highly accurate method of regressing parameters from medical imaging data that does not rely on hand-selected features. They may be more readily extended to new applications than traditional machine learning techniques.
Notes
- 1.
Performed by an expert in the Tschannen et al. [9] group.
- 2.
When tested during development tanh produced a better loss than ReLU when used in these stages. Possibly due to the fact that tanh does not force the output to positive numbers.
- 3.
Foregoing the activation function allows this stage to produce the full range of floats as possible outputs.
References
Kim, S., Wise, B., Zhang, Y., Szabo, R.: Increasing incidence of shoulder arthroplasty in the United States. J. Bone Joint Surg. Am. 93(24), 2249–2254 (2011). https://doi.org/10.2106/JBJS.J.01994
Keener, J., Chalmers, P., Yamaguchi, K.: The humeral implant in shoulder arthroplasty. J. Am. Acad. Orthop. Surg. 25(6), 427–438 (2017). https://doi.org/10.5435/JAAOS-D-15-00682
Edwards, T., Morris, B., Gartsman, G.: Shoulder Arthroplasty, 2nd edn. Elsevier, Amsterdam (2019)
Dines, D., Laurencin, C., Williams, G. (eds.): Arthritis & Arthroplasty: The Shoulder. Saunders/Elsevier, Philadelphia (2009)
Pearl, M.: Proximal humeral anatomy in shoulder arthroplasty: implications for prosthetic design and surgical technique. J. Shoulder Elbow Surg. 14(Suppl 1), S99–S104 (2005). https://doi.org/10.1016/j.jse.2004.09.025
DeLude, J., et al.: An anthropometric study of the bilateral anatomy of the humerus. J. Shoulder Elbow Surg. 16(4), 477–483 (2007). https://doi.org/10.1016/j.jse.2006.09.016
Johnson, J., Thostenson, J., Suva, L., Hasan, S.: Relationship of bicipital groove rotation with humeral head retroversion: a three-dimensional computed tomographic analysis. J. Bone Joint Surg. Am. 95(8), 719–724 (2013). https://doi.org/10.2106/JBJS.J.00085
Vlachopoulos, L., et al.: Computer algorithms for three-dimensional measurement of humeral anatomy: analysis of 140 paired humeri. J. Shoulder Elbow Surg. 25(2), e38–e48 (2016). https://doi.org/10.1016/j.jse.2015.07.027
Tschannen, M., Vlachopoulos, L., Gerber, C., Székely, G., Fürnstahl, P.: Regression forest-based automatic estimation of the articular margin plane for shoulder prosthesis planning. Med. Image Anal. 31, 88–97 (2016). https://doi.org/10.1016/j.media.2016.02.008
Janssens, R., Zeng, G., Zheng, G.: Fully automatic segmentation of lumbar vertebrae from CT images using cascaded 3D fully convolutional networks. arXiv:1712.01509 (2017). http://arxiv.org/abs/1712.01509
Payer, C., Štern, D., Bischof, H., Urschler, M.: Regressing heatmaps for multiple landmark localization using CNNs. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 230–238. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_27
Zhang, J., et al.: Joint craniomaxillofacial bone segmentation and landmark digitization by context-guided fully convolutional networks. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D., Duchesne, S. (eds.) MICCAI 2017. LNCS, vol. 10434, pp. 720–728. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66185-8_81
Boileau, P., Cheval, D., Gauci, M., Holzer, N., Chaoui, J., Walch, G.: Automated three-dimensional measurement of glenoid version and inclination in arthritic shoulders. J. Bone Joint Surg. Am. 100(1), 57–65 (2018). https://doi.org/10.2106/JBJS.16.01122
Nguyen, D., et al.: Improved accuracy of computer assisted glenoid implantation in total shoulder arthroplasty: an in-vitro randomized controlled trial. J. Shoulder Elbow Surg. 18(6), 907–914 (2009). https://doi.org/10.1016/j.jse.2009.02.022
Werner, B., Hudek, R., Burkhart, K., Gohlke, F.: The influence of three-dimensional planning on decision-making in total shoulder arthroplasty. J. Shoulder Elbow Surg. 26(8), 1477–1483 (2017). https://doi.org/10.1016/j.jse.2017.01.006
Suzani, A., Seitel, A., Liu, Y., Fels, S., Rohling, R.N., Abolmaesumi, P.: Fast automatic vertebrae detection and localization in pathological CT scans - a deep learning approach. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 678–686. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_81
Kingma, D., Ba, J.: Adam: a method for stochastic optimization. arXiv:1412.6980 (2014). http://arxiv.org/abs/1412.6980
Abadi, M., et al.: TensorFlow: a system for large-scale machine learning. arXiv:1605.08695 (2016). http://arxiv.org/abs/1605.08695
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Kulyk, P., Vlachopoulos, L., Fürnstahl, P., Zheng, G. (2019). Fully Automatic Planning of Total Shoulder Arthroplasty Without Segmentation: A Deep Learning Based Approach. In: Vrtovec, T., Yao, J., Zheng, G., Pozo, J. (eds) Computational Methods and Clinical Applications in Musculoskeletal Imaging. MSKI 2018. Lecture Notes in Computer Science(), vol 11404. Springer, Cham. https://doi.org/10.1007/978-3-030-11166-3_3
Download citation
DOI: https://doi.org/10.1007/978-3-030-11166-3_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-11165-6
Online ISBN: 978-3-030-11166-3
eBook Packages: Computer ScienceComputer Science (R0)