Abstract
Magnetic resonance imaging (MRI) is crucial for in vivo detection and characterization of white matter lesions (WML) in multiple sclerosis (MS). The most widely established MRI outcome measure is the volume of hyperintense lesions on T2-weighted images (T2L). Unfortunately, T2L are non-specific for the level of tissue destruction and show a weak relationship to clinical status. Interest in lesions appearing hypointense on T1-weighted images (T1L) (“black holes”), which provide more specificity for axonal loss and a closer link to neurologic disability, has thus grown. The technical difficulty of T1L segmentation has led investigators to rely on time-consuming manual assessments prone to inter- and intra-rater variability. We implement MIMoSA, a current T2L automatic segmentation approach, to delineate T1L. Using cross-validation, MIMoSA proved robust for segmenting both T2L and T1L. For T2L, a Sørensen-Dice coefficient (DSC) of 0.6 and partial AUC (pAUC) up to 1% false positive rate of 0.69 were achieved. For T1L, 0.48 DSC and 0.63 pAUC were achieved. The correlation between EDSS and manual versus automatic volumes were similar for T1L (0.32 manual vs. 0.34 MIMoSA) and T2L (0.34 vs. 0.34).
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Multiple sclerosis (MS) is a life-long chronic disease of the central nervous system with no known cure. MS is the most common autoimmune disorder globally with about 2.3 million people affected worldwide [1, 2]. The pathophysiology of MS includes development of lesions which occur in the white matter (WML) and exhibit inflammation, destruction of myelin sheaths, and axonal loss. The accumulation of WML is associated with long-term morbidity and disability and is visible on structural magnetic resonance imaging (MRI).
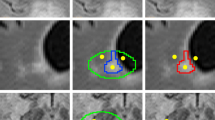
Common quantitative MRI metrics in MS include lesion volume and count which rely on accurate segmentation of WML. Lesion count and volume are often derived from three related pathological presentations of WML: (1) contrast-enhancing lesions (EL), which are thought to represent acute perivascular inflammatory activity following focal break-down of the blood brain barrier, (2) T2 hyperintense lesions (T2L), which detect the process of demyelination and axonal loss and non-specific damage unrelated to MS, and (3) persisting T1 hypointense lesions (T1L), which are the most demyelinated and damaged regions [3,4,5]. Figure 1 displays axial slices of FLAIR and T1-weighted images with manual delineations overlaid.
Despite the existence of a number of automatic lesion segmentation methods [6,7,8,9,10], the majority delineate T2L solely. The sparsity of prior research is in part due to technical challenges: since T1L and their boundaries appear similar to gray matter [11] and are subtler than T2L, they are much more difficult to segment by manual and automatic methods. The simplest method was proposed by Filippi et al. using an expert-driven semi-automated thresholding approach to estimate lesion volumes [12]. Molyneux et al. similarly propose a semi-automated technique to delineate T1L in a multi-center study where they showed that T1L volume is a consistent and reproducible metric that can be applied to MRI data from various scanners [13]. Following these results, Datta et al. recently developed fully automated methods using fuzzy connectivity modeling [14]. Other methods proposed an algorithm to detect EL, T1L, and T2L using intensity-based statistical k-nearest neighbor classification combined with template-driven segmentation and partial volume artifact correction [15]. To automatically segment T1L, others proposed an approach that used a standard classification algorithm to partition T1-weighted images into gray matter, white matter, and cerebrospinal fluid and then finding T1L in the white matter by spatially voxel-wise testing using healthy controls as a reference [16]. Unfortunately, no approach has released publicly available software and these studies were based on relatively small MRI data sets with uniform patient demographics and lesion load [12,13,14,15,16].
In our previous work, a Method for Inter-Modal Segmentation Analysis (MIMoSA) was developed and validated as an automatic T2L segmentation method in people with MS [10]. In the present study, we extended the MIMoSA method to automatically segment T1L. Since no publicly available software for automatic detection of T1L exists, we automatically segment T2L using MIMoSA and used these measures as a reference for T1L performance. This was motivated by our findings that MIMoSA is a competitive T2L segmentation approach [10], and all T1L are also seen as T2L (but not vice-versa). Moreover, since the data acquired in this study were acquired under different protocol than data in the original development of MIMoSA, through the application of MIMoSA to segment T2L we validate and assess robustness of MIMoSA’s accuracy across scanner platforms and protocols. For further comparison, OASIS, another validated T2L lesion segmentation algorithm [8], was used to automatically segment T1L. Finally, we examined correlations between lesion volume with clinical status measurements in order to determine if the reduction in noise associated with automatic lesion segmentation revealed stronger associations with disability.
2 Methods
2.1 Data and Preprocessing
Data were collected at the Brigham and Women’s Hospital in Boston, Massachusetts. Forty patients, all with a clinical diagnosis of MS, were consecutively obtained from MRI scans at the center. Subjects had an examination by an MS specialist neurologist to assess the type of MS, the level of physical disability on the Expanded Disability Status Scale (EDSS), and ambulatory function on the timed 25-foot walk (T25FW). High-resolution 3D T1-weighted (T1WI), T2-weighted (T2WI), and fluid-attenuated inversion recovery (FLAIR) volumes of the brain were collected on a Siemens 3T Skyra instrument using a consistent scan protocol among subjects. In addition to the imaging sequences, T1L and T2L were manually segmented by an experienced reading panel of two observers under the supervision of an experienced observer. The observers determined the presence or absence of T1L together to form a single consensus segmentation between the two raters. In the event of a disagreement, a senior experienced observer was consulted. This procedure was repeated to segment T2L so that T1L and T2L were segmented by the two raters using a consensus approach but the lesion types are obtained independently. These T1L and T2L manual annotations were acquired manually and without the use of any automatic method.
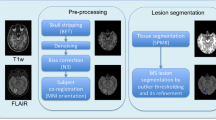
All images were preprocessed prior to implementing the MIMoSA model, using the R (version 3.1.0, R Foundation for Statistical Computing, Vienna, Austria) packages extrantsr [17] and WhiteStripe [18] as well as Multi-Atlas Skull-Stripping (MASS) [19, 20]. After N4 inhomogeneity correction [21], volumes were co-registered across sequences for each subject using a rigid-body transformation with a Lanczos windowed sinc interpolator. To remove extracerebral voxels, MASS was implemented [19, 20]. As conventional MRI volumes are acquired in arbitrary units, statistical intensity normalization using WhiteStripe [18] was applied in order to model intensities across subjects.
2.2 MIMoSA: Method for Inter-modal Segmentation Analysis
MIMoSA was originally built to automatically segment T2L and extended in this pilot study to automatically segment T1L. As feature extraction is known to be pivotal for a segmentation algorithm’s accuracy and generalizability, the MIMoSA method relies on features that capture the mean structure of each imaging modality as well as the covariance across modalities. The method requires FLAIR and T1 as image inputs but can also utilize T2 and PD if they were collected. The MIMoSA procedure is outlined in Fig. 2 and described below. First, MIMoSA identifies all candidate voxels for T2L presence defined as 85th percentile or above on the FLAIR. The algorithm then creates the features to be implemented in the model. The features included in the model are normalized MRI volumes, smoothed volumes with kernel parameters 10 and 20 mm, and inter-modal coupling (IMCo) intercept and slope coefficients for each combination of images as outcome and predictors [22]. With all relevant features calculated, the procedure then fits a local logistic regression based on gold standard manual segmentations to obtain coefficients which we then use to produce maps of the probability of lesion.
In the model below, \( P\left\{ {L_{i} \left( v \right) = 1} \right\} \) represents the probability that a voxel is part of a lesion where \( L_{i} \left( v \right) \) is a random variable denoting voxel-level lesion presence. If there is a lesion in voxel \( v \) for subject \( i \), then \( L_{i} \left( v \right) = 1 \), otherwise \( L_{i} \left( v \right) = 0 \). We model the probability that a voxel \( v \) contains lesion incidence with the following logistic regression model:
where we denote the normalized images \( X_{i} \left( v \right) = \left[ {T_{1,i} \left( v \right), FLAIR_{i} \left( v \right), T_{2,i} \left( v \right),PD_{i} \left( v \right)} \right]^{T} \) and we use \( {\mathfrak{G}} \) to denote the smoothing operator with parameter \( \delta \in \left\{ {10\,{\text{mm}},20\,{\text{mm}}} \right\} \), which takes a weighted average within each neighborhood \( N\left( {v,\delta } \right) \) around \( v \). We express the smoothed images in vector form by \( {\mathfrak{G}}X_{i} \left( {v,\delta } \right) = \left[ {{\mathfrak{G}}\left( {T_{1,i} \left( v \right);N\left( {v,\delta } \right)} \right), \ldots ,{\mathfrak{G}}\left( {PD_{i} \left( v \right);N\left( {v,\delta } \right)} \right)} \right]^{T} \), and we denote all combination of intercept and slope IMCo parameters respectively by \( {\mathcal{C}}X_{i,I}^{T} \left( v \right) \) and \( { \mathcal{C}}X_{i,S}^{T} \left( v \right) \). We use \( \otimes \) to represent the Hadamard product. The interaction terms between the normalized volumes and the smoothed volumes, denoted by \( \beta_{j0}^{*} \), contribute to the model by capturing differences between voxel intensities and their local mean intensities. These aid in mitigating artifacts due to residual field inhomogeneity in some cases, and generally improve lesion detection performance. We use a logistic regression because it is simple, easy to interpret, and computationally quick [8]. In the past, studies have compared classification methods and shown that simple methods often yield performance equivalent to more sophisticated methods so long as relevant biological features are included [23].
After fitting, the MIMoSA method can then be applied to new subjects, namely subjects not included in the training set, in order generate probability maps which we then threshold to create binary lesion segmentation masks. We select the threshold by an optimal thresholding algorithm that optimizes similarity of predicted segmentation masks in the training set with gold standard segmentations based on DSC. To automatically segment T2L and T1L separate models must be fit based on manual segmentations. We simply apply the MIMoSA procedure, built for T2L to T1L.
2.3 Statistical Analysis
Training and testing of MIMoSA methods was conducted using a cross-validation. In addition to implementing MIMoSA, a competitive T2L segmentation algorithm, OASIS, was also applied [8]. OASIS was specifically chosen for the present study as it can be easily trained using publicly available software and there are no publicly available data for benchmarking T1L automatic lesion segmentation. To fit the models and measure performance, 20 subjects were allocated to the training set and 20 subjects to the test set. MIMoSA and OASIS were then trained for T1L and T2L separately using subjects in the training set. After models were fit, the estimated coefficients were applied to the test set in order to generate probability maps. To generate lesion masks, the threshold produced from the optimal threshold algorithm described above was applied.
This procedure was iterated 100 times. In each fold, subject-level DSC and partial AUC (pAUC, up to 1% false positive rate) were recorded [24]. pAUC was estimated rather than traditional AUC since it only considers regions of the ROC space which correspond to clinically relevant values of specificity [25]. After calculation at the subject level, performance measures were averaged across subjects and cross-validation folds. Figure 3 shows the full cross-validation pipeline.
Bootstrap cross-validation scheme to assess MIMoSA performance on T1 lesion (T1L) and T2 lesion (T2L). To identify the optimal threshold, probability maps for subjects in the training set were generated using the model previously fit. These maps were thresholded along a grid and Sørensen-Dice coefficient (DSC) was calculated. The threshold resulting in the maximum DSC across subjects in the training set was applied to the threshold in the test set.
3 Results
The DSC and pAUC false positive rates up to 1% are shown in Fig. 4. False positive rates above 1% are not clinically useful in MS lesion segmentation. For example, consider 1% of the volume of a healthy control subject is on the order of 10 cm3, which is equal to the average lesion load of an MS subject. Results in Fig. 4 indicate competitive lesion segmentation performance of both T1L and T2L. The method accurately delineates T1L and T2L as exemplified by high DSC and pAUC. MIMoSA performance measures are all higher than OASIS results indicating superior automatic segmentation.
Results from the cross-validation are presented. T1 lesion (T1L) and T2 lesion (T2L) average measures for Sørensen-Dice coefficient (DSC) and partial AUC (pAUC) with up to 1% false positive rate were averaged within each testing set and then across folds. Error bars are overlaid where standard deviation was calculated within cross-validation folds and averaged iterations.
In practice, common applications of lesion segmentation metrics are for association studies with clinical status and evaluating therapeutic efficacy [5, 26]. In Table 1, we report the relationship between both manual and MIMoSA lesion segmentation metrics and clinical measures. Volumetric measures were correlated with EDSS score and T25FW for T1L and T2L. The correlations displayed in this table show that \( \hat{\rho }\left( {MIMoSA} \right) \) is equal to or larger than \( \hat{\rho }\left( {Manual} \right) \).
Lesion volume and count are important metrics for diagnosis and in the evaluation of therapeutic effectiveness. Thus, their accurate estimation from an automatic method is of the utmost importance. Figure 5 provides subject-level measures of volume and count using MIMoSA compared with manually acquired metrics, averaged across cross validation iterations. MIMoSA’s estimation of lesion volume is extremely accurate as the T1L and T2L points all lie close to the \( {\text{y}} = {\text{x}} \) line. Additionally, the correlations presented overlaid on the graph are very close to 1. Lesion count is similarly very accurate for subjects with less than 25 lesions. As lesion count increases beyond this though, MIMoSA tends to undercount lesions.
4 Discussion
MIMoSA is a fully automated segmentation method that utilizes changes in inter-modal covariance structure occurring in white matter pathology, and can be used to delineate T1L and T2L accurately, reliably, and efficiently in people with MS. The use of IMCo likely drives improvements in accuracy as IMCo measures appear to be robust to even small changes in intensity across imaging modalities. These measures are especially useful for detecting T1L, a challenging task since these lesions appear similar to gray matter. MIMoSA does not require human input, providing stability and consistency into delineation. The model can easily be adapted and trained for cases with more or fewer imaging sequences [8, 10]. The full modeling procedure can be easily and quickly implemented using software and documentation provided through Neuroconductor [27, 28].
MIMoSA provides accurate and reliable automatic segmentations of both T1L and T2L. Though T2L DSC and pAUC measures are slightly larger, indicating more similarity with manual segmentations, T1L performance was competitive. The automatic segmentation of T1L and T2L using the same procedure allows for a simple and consistent framework to obtain both metrics. Simultaneous delineation of T1L and T2L will lead to a better understanding of overall patient status. The correlation between manual volumes and MIMoSA volumes of WML indicates precision for total volume prediction. Since total lesion volume is commonly used in the assessment of new therapies in clinical trials [5], MIMoSA provides a promising alternative to manual segmentation in these settings. This may be especially useful for multi-center studies with a large number of patients or longitudinal studies with sequences collected over time.
Often lesion volumes are correlated with clinical covariates and disease status in patient management and clinical trials that evaluate therapy effectiveness. Therefore, automatic segmentation approaches should be as sensitive as manual measures. Correlations were provided to compare manual and MIMoSA segmentations with clinically relevant variables. Our results indicate that the relationship between MIMoSA volumetric assessments showed as close or better correlations compared with using manual segmentations. This was likely due to the stability and consistency introduced by an automatic method that requires no operator input. Segmentation of T1L can be challenging since the intensity profile is often indistinguishable from gray matter [5], especially with respect to delineating boundaries; thus, reliability in these areas may be the cause for the stronger correlation with covariates. For T2L evaluation, correlations seem to be approximately equal between MIMoSA and manual segmentations. In general, the measurements, whether obtained from manual segmentation or MIMoSA, were similar, advocating for the use of the automated method to cut cost, time, and introduce stability without sacrificing relation to patient status.
References
Berer, K., Krishnamoorthy, G.: Microbial view of central nervous system autoimmunity. FEBS Lett. 588, 4207–4213 (2014)
Feigin, V.L., et al.: Global, regional, and national burden of neurological disorders during 1990–2015: a systematic analysis for the global burden of disease study 2015. Lancet Neurol. 16, 877–897 (2017)
Rovira, À., León, A.: MR in the diagnosis and monitoring of multiple sclerosis: an overview. Eur. J. Radiol. 67, 409–414 (2008)
Katdare, A., Ursekar, M.: Systematic imaging review: multiple sclerosis. Ann. Indian Acad. Neurol. 18, S24–S29 (2015)
Bakshi, R., Minagar, A., Jaisani, Z., Wolinsky, J.S.: Imaging of multiple sclerosis: role in neurotherapeutics. NeuroRx 2, 277–303 (2005)
García-Lorenzo, D., Francis, S., Narayanan, S., Arnold, D.L., Collins, D.L.: Review of automatic segmentation methods of multiple sclerosis white matter lesions on conventional magnetic resonance imaging. Med. Image Anal. 17, 1–18 (2013)
Meier, D.S., et al.: Dual-sensitivity multiple sclerosis lesion and CSF segmentation for multichannel 3T brain MRI. J. Neuroimaging Off. J. Am. Soc. Neuroimaging 28, 36–47 (2017)
Sweeney, E.M., et al.: OASIS is automated statistical inference for segmentation, with applications to multiple sclerosis lesion segmentation in MRI. NeuroImage Clin. 2, 402–413 (2013)
Shiee, N., Bazin, P.-L., Ozturk, A., Reich, D.S., Calabresi, P.A., Pham, D.L.: A topology-preserving approach to the segmentation of brain images with multiple sclerosis lesions. NeuroImage 49, 1524–1535 (2010)
Valcarcel, A.M., et al.: MIMoSA: an automated method for intermodal segmentation analysis of multiple sclerosis brain lesions. J. Neuroimaging 28, 389–398 (2018)
Ceccarelli, A., et al.: The impact of lesion in-painting and registration methods on voxel-based morphometry in detecting regional cerebral gray matter atrophy in multiple sclerosis. AJNR Am. J. Neuroradiol. 33, 1579–1585 (2012). PubMed – NCBI. https://www.ncbi.nlm.nih.gov/pubmed/22460341
Filippi, M., Rovaris, M., Campi, A., Pereira, C., Comi, G.: Semi-automated thresholding technique for measuring lesion volumes in multiple sclerosis: effects of the change of the threshold on the computed lesion loads. Acta Neurol. Scand. 93, 30–34 (1996)
Molyneux, P.D., et al.: The precision of T1 hypointense lesion volume quantification in multiple sclerosis treatment trials: a multicenter study. Mult. Scler. J. 6, 237–240 (2000)
Datta, S., Sajja, B.R., He, R., Wolinsky, J.S., Gupta, R.K., Narayana, P.A.: Segmentation and quantification of black holes in multiple sclerosis. NeuroImage 29, 467–474 (2006)
Wu, Y., et al.: Automated segmentation of multiple sclerosis lesion subtypes with multichannel MRI. NeuroImage 32, 1205–1215 (2006)
Spies, L., et al.: Fully automatic detection of deep white matter T1 hypointense lesions in multiple sclerosis. Phys. Med. Biol. 58, 8323–8337 (2013)
Muschelli, J.: extrantsr: extra functions to build on the ANTsR package (2014)
Shinohara, R.T., Muschelli, J.: WhiteStripe: white matter normalization for magnetic resonance images using WhiteStripe (2017)
Doshi, J., Erus, G., Ou, Y., Gaonkar, B., Davatzikos, C.: Multi-Atlas Skull-Stripping. Acad. Radiol. 20, 1566–1576 (2013)
NITRC: CBICA: Multi Atlas Skull Stripping (MASS): Tool/Resource Info. https://www.nitrc.org/projects/cbica_mass/
Tustison, N.J., et al.: N4ITK: improved N3 bias correction. IEEE Trans. Med. Imaging 29, 1310–1320 (2010)
Vandekar, S.N.: Subject-level measurement of local cortical coupling. NeuroImage 133, 88–97 (2016)
Hand, D.J., et al.: Classifier technology and the illusion of progress. Stat. Sci. 21, 1–14 (2006)
Sing, T., Sander, O., Beerenwinkel, N., Lengauer, T.: ROCR: visualizing classifier performance in R. Bioinformatics 21, 3940–3941 (2005)
Walter, S.D.: The partial area under the summary ROC curve. Stat. Med. 24, 2025–2040 (2005)
Zivadinov, R., Bakshi, R.: Role of MRI in multiple sclerosis I: inflammation and lesions. Front. Biosci. J. Virtual Libr. 9, 665–683 (2004)
Neuroconductor. https://neuroconductor.org/
Mimosa—Neuroconductor. https://neuroconductor.org/package/details/mimosa
Acknowledgements
This project was supported in part by a pilot grant from the Center for Biomedical Computing and Analytics at the University of Pennsylvania as well as R01NS085211, R21NS093349, R01NS060910, and R01MH112847 from the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Valcarcel, A.M. et al. (2019). MIMoSA: An Approach to Automatically Segment T2 Hyperintense and T1 Hypointense Lesions in Multiple Sclerosis. In: Crimi, A., Bakas, S., Kuijf, H., Keyvan, F., Reyes, M., van Walsum, T. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2018. Lecture Notes in Computer Science(), vol 11383. Springer, Cham. https://doi.org/10.1007/978-3-030-11723-8_5
Download citation
DOI: https://doi.org/10.1007/978-3-030-11723-8_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-11722-1
Online ISBN: 978-3-030-11723-8
eBook Packages: Computer ScienceComputer Science (R0)