Abstract
Brain tumor is one of the leading causes of cancer death. Accurate segmentation and quantitative analysis of brain tumor are critical for diagnosis and treatment planning. Since manual segmentation is time-consuming, tedious and error-prone, a fully automatic method for brain tumor segmentation is needed. Recently, state-of-the-art approaches for brain tumor segmentation are built on fully convolutional neural networks (FCNs) using either 2D or 3D convolutions. However, 2D convolutions cannot make full use of the spatial information of volumetric medical image data, while 3D convolutions suffer from high expensive computational cost and memory demand. To address these problems, we propose a novel Separable 3D U-Net architecture using separable 3D convolutions. Preliminary results on BraTS 2018 validation set show that our proposed method achieved a mean enhancing tumor, whole tumor, and tumor core Dice scores of 0.74932, 0.89353 and 0.83093 respectively. Finally, during the testing stage we achieved competitive results with Dice scores of 0.68946, 0.83893, and 0.78347 for enhancing tumor, whole tumor, and tumor core, respectively.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Image segmentation, especially semantic segmentation, is a fundamental and classic problem in computer vision. It refers to partitioning an image into several disjoint semantically meaningful parts and classifying each part into a pre-determined class. In the application of brain tumor segmentation, the task includes the division of several sub-regions, such as GD-enhancing tumor, peritumoral edema, and the necrotic and non-enhancing tumor core [1]. Accurate segmentation and quantitative analysis of brain tumor are critical for diagnosis and treatment planning. Generally, manual segmentation of brain tumor is known to be time-consuming, tedious and error-prone. Therefore, there is a strong need for a fully automatic method for brain tumor segmentation. However, brain tumor segmentation is a challenging task because MR images are typically acquired using various protocols and magnet strengths, which results in the non-standard range of MR images. In addition, brain tumors can appear anywhere in the brain, and their shape and size vary greatly. Furthermore, the intensity profiles of tumor regions are largely overlapped with healthy parts. Due to the challenge of brain tumor segmentation and the broad medical prospect, many researchers have proposed various methods to solve the problem of brain tumor segmentation.
Brain tumor segmentation methods can be divided into different categories according to different principles [2]. Broadly, these methods can be divided into two major categories: generative methods and discriminative methods. Generative methods typically rely on the prior information about the appearance of both healthy tissues and tumors. The proposed models often regard the task of segmentation as a problem of a posteriori distribution estimation. On the contrary, discriminative methods use very little prior information and typically rely on a large number of low-level image features to learn the distribution from the annotated training images.
More recently, due to the success of convolutional neural networks (CNNs), great progress has been made in the field of computer vision. At the same time, many deep learning based brain tumor segmentation methods have been proposed and achieved great success. Havaei et al. [3] proposed a two-pathway architecture with a local pathway and a global pathway, which can simultaneously exploit both local features and more global contextual features. Kamnitsas et al. [4] proposed an efficient fully connected multi-scale CNN architecture named deepmedic that uses 3D convolution kernels and reassembles a high resolution and a low resolution pathway to obtain the segmentation results. Furthermore, they used a 3D fully connected conditional random field to effectively remove false positives. Isensee et al. [5] proposed 3D U-Net, which carefully modified the popular U-Net architecture and used a dice loss function to cope with class imbalance. They achieved competitive results on the BraTS 2017 testing data. Kamnitsas et al. [6] introduced EMMA, an ensemble of multiple models and architectures including deepmedic, FCNs and U-Net. Due to the heterogeneous collection of networks, the model is insensitive to independent failures of each component and has good generalization performance. They won first place in the final testing stage of the BraTS 2017 challenge among more than 50 teams.
Although so many achievements have been made, the progress of medical image analysis is slower than that of static images, and a key reason is the 3D properties of medical images. This problem also occurs in the tasks of video understanding. To solve this problem, Xie et al. [7] proposed S3D model by replacing 3D convolutions with spatiotemporal-separable 3D convolutions. This model significantly improved on the previous state-of-the-art 3D CNN model in terms of efficiency.
Inspired by S3D architecture for video classification and the state-of-the-art U-Net architecture for medical image segmentation, we propose a novel framework named S3D-UNet for brain tumor segmentation. To make full use of 3D volumes, we design a new separable 3D convolution by dividing each 3D convolution into three branches in a parallel fashion, each with a different orthogonal view, namely axial, sagittal and coronal. We also propose a separable 3D block that takes advantage of the state-of-the-art residual inception architecture. During the testing stage we achieved competitive results with Dice scores of 0.68946, 0.83893, and 0.78347 for enhancing tumor, whole tumor, and tumor core, respectively [8].
2 Methods
2.1 Dataset
The brain tumor MRI dataset used in this study are provided by BraTS’2018 Challenge [1, 9,10,11]. The training dataset includes multimodal brain MRI scans of 285 subjects, of which 210 are GBM/HGG and 75 are LGG. Each subject contains four scans: native T1-weighted (T1), post-contrast T1-weighted (T1c), T2-weighted (T2), and T2 Fluid Attenuated Inversion Recovery (FLAIR). All the subjects in the training dataset are provided with ground truth labels, which are segmented manually by one to four raters. Annotations consist of the GD-enhancing tumor (ET - label 4), the peritumoral edema (ED - label 2), and the necrotic and non-enhancing tumor core (NCR/NET - label 1). The validation and testing datasets include multimodal brain MRI scans of 66 subjects and 191 subjects which are similar to the training dataset but have no expert segmentation annotations and the grading information.
2.2 Data Pre-processing
To remove the bias field caused by the inhomogeneity of the magnetic field and the small motions during scanning, the N4ITK bias correction algorithm [12] is first applied to the T1, T1c and T2 scans. The multimodal scans in BraTS 2018 were acquired with different clinical protocols and various scanners from multiple institutions [1], resulting in non-standardized intensity distribution. Therefore, normalization is a necessary stage of processing multi-mode scanning by a single algorithm. We use the histogram matching algorithm [13] to transform each scan to a specified histogram to ensure that all the scans have a similar intensity distribution. We also resize the original image of \( 240 \times 240 \times 155 \) voxels to \( 128 \times 128 \times 128 \) voxels by removing as many zero background as possible. This processing not only can effectively improve the calculation efficiency, but also retain the original image information as much as possible. In the end, we normalize the data to have a zero mean and unit variance.
2.3 Network Architecture
S-3D Convolution Block. Traditional 2D CNNs for computer vision mainly involve spatial convolutions. However, for video applications such as human action, both spatial and temporal information need to be modeled jointly. By using 3D convolution in the convolutional layers of CNNs, discriminative features along both the spatial and the temporal dimensions can be captured. 3D CNNs have been widely used for human action recognition in videos. However, the training of 3D CNN requires expensive computational cost and memory demand, which hinders the construction of a very deep 3D CNN. To mitigate this problem, Xie et al. [7] proposed S3D model by replacing 3D convolutions with spatiotemporal-separable 3D convolutions. Each 3D convolution can be replaced by two consecutive convolutional layers: one 2D convolution to learn spatial features and one 1D convolution to learn temporal features, as shown in Fig. 1(a). By using separable temporal convolution, they build a new block using inception architecture called “temporal inception block”, as shown in Fig. 1(b).
Unlike video data, volumetric medical data have three orthogonal views, namely axial, sagittal and coronal, and each view has important anatomical features. To implement the separable 3D convolution directly, we need to specify which view as the temporal direction. Wang et al. [14] propose a cascaded anisotropic convolutional neural network consisting of multiple layers of anisotropic convolution filters, which are then combined with multi-view fusion to reduce false positives. Each view of this architecture is similar to a separable 3D convolution, and the multi-view fusion can be view as an ensemble of networks in three orthogonal views that utilize 3D contextual information for higher accuracy. They train a neural network for each view, it is not end-to-end and requires longer time for training and testing. To fully utilize 3D contextual information and reduce computational complexity, we divide a 3D convolution into three branches in a parallel fashion, each with a different orthogonal view, as shown in Fig. 2(a). Furthermore, we propose a separable 3D block that takes advantage of the residual inception architecture, as shown in Fig. 2(b).
S3D U-Net Architecture.
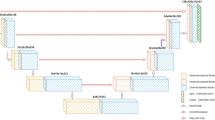
Our framework is based on the U-Net structure proposed by Ronneberger et al. [15] which consists of a contracting path to analyze the whole image and a symmetric expanding path to recovery the original resolution, as shown in Fig. 3. The U-Net structure has been widely used in the field of medical image segmentation and has achieved competitive performance. Several studies [5, 16] have demonstrated that a 3D version of U-Net using 3D volumes as input can produce better results than an entirely 2D architecture.
Just like the U-Net and its extensions, our network has an autoencoder-like architecture with a contracting path and an expanding path, as shown in Fig. 3. The contracting path encodes the increasingly abstract representation of the input, and the expanding path restores the original resolution. Similar to [5], we refer to the depth of the network as level. Higher levels have lower spatial resolution but higher dimensional feature representations and vice versa.
The input to the contracting path is a \( 128 \times 128 \times 128 \) voxel block with 4 channels. The contracting path has 5 levels. Except for the first level, each level consists of two S3D blocks. It is worth noting that each convolution in S3D block is followed by instance normalization [17] and LeakyReLU. Different levels are connected by transition down block to reduce the resolution of the feature maps and double the number of feature channels. Transition down module consists of a \( 3 \times 3 \times 3 \) convolution with stride 2 followed by instance normalization and LeakyReLU. After the contracting path, the size of the feature maps is decreased to \( 8 \times 8 \times 8 \).
In order to recover the input resolution at expanding path, we first adopt a transition up module to upsample the previous feature maps and halve the number of feature channels. Transition up module consists of a transposed \( 3 \times 3 \times 3 \) convolution with stride 2 followed by instance normalization and LeakyReLU. Then the feature maps from contracting path are concatenated with feature maps from expanding path via long skip connections. At each level of expanding path, we use a \( 1 \times 1 \times 1 \) convolution with stride 1 to halve the number of feature channels, followed by two S3D blocks that are the same as in the contracting path. The final segmentation is done by a \( 1 \times 1 \times 1 \) convolutional layer followed by a softmax operation among the objective classes.
2.4 Loss Function
The performance of neural network depends not only on the choice of network structure but also on the choice of the loss function [18]. Especially for severe class imbalance, the choice of loss functions becomes more important. Due to the physiological characteristics of brain tumors, the segmentation task has an inherent class imbalance problem. Table 1 illustrates the distribution of the classes in the training data of BraTS 2018. Background (label 0) is overwhelmingly dominant. According to [5], we apply a multiclass Dice loss function to approach this issue. Let \( R \) be the one hot coding ground truth segmentation with voxel values \( r_{n}^{k} \), where \( k \in K \) being the class at voxel \( n \in N \). Let \( P \) be the output the network with voxel values \( p_{n}^{k} \), where \( k \in K \) being the class at voxel \( n \in N \). The multiclass Dice loss function can be expressed as
2.5 Evaluation Metrics
Multiple criteria are computed as performance metrics to quantify the segmentation result. Dice coefficient (Eq. 2) is the most frequently used metric for evaluating medical image segmentation. \( P_{1} \) is the area that is predicted to be tumor and \( T_{1} \) is true tumor area. It measures the overlap between the segmentations and ground truth with a value between 0 and 1. The higher the Dice score, the better the segmentation performance.
Sensitivity and specificity are also commonly used statistical measures. The sensitivity (Eq. 3), also called true positive rate, defined as the proportion of positives that are correctly predicted. It measures the portion of tumor regions in the ground truth that are also predicted as tumor regions by the segmentation method. The specificity (Eq. 4), also called true negative rate, defined as the proportion of negatives that are correctly predicted. It measures the portion of normal tissue regions \( \left( {T_{0} } \right) \) in the ground truth that are also predicted as normal tissue regions \( \left( {P_{0} } \right) \) by the segmentation method.
The Hausdorff Distance (Eq. 5) is used to evaluates the distance between the segmentation boundary and the ground truth boundary. Mathematically, it is defined as the maximum distance of all points \( p \) on the surface \( \partial P_{1} \) of a given volume \( P_{1} \) to the nearest points \( t \) on the surface \( \partial T_{1} \) of the other given volume \( T_{1} \).
3 Experiments and Results
The network is trained on a GeForce GTX 1080Ti GPU with a batch size of 1 using PyTorch toolbox. Adam [19] is used as the optimizer with an initial learning rate 0.001 and a l2 weight decay of 1e−8. We evaluate all the cases for training data and validation data using online CBICA portal for BraTS 2018 challenge. The sub-regions considered for evaluation are “enhancing tumor” (ET), “tumor core” (TC), and “whole tumor” (WT).
Table 2 presents the quantitative evaluations with the BraTS 2018 training set via five cross-validation. It shows that the proposed method achieves average Dice scores of 0.73953, 0.88809 and 0.84419 for enhancing tumor, whole tumor and tumor core, respectively. A 3D U-Net without the proposed S3D block is also trained, and the quantitative evaluations with the BraTS 2018 training set are shown in Table 3. It can be seen that the Dice score of enhancing tumor has been significantly improved using S3D block. The corresponding values for BraTS 2018 validation set are 0.74932, 0.89353 and 0.83093, respectively, as shown in Table 4. Examples of the segmentations obtained from the training set using our method are shown in Fig. 4.
Examples of segmentation from the of BraTS 2018 training data. red: NCR/NET, green: ED, blue: ET. (the first two rows) Satisfying segmentation. (the last two rows) Unsatisfactory segmentation. In the future, we will adopt some post-processing methods to improve the segmentation performance. (Color figure online)
Table 5 shows the challenge testing set results. Our proposed method achieves average Dice scores of 0.68946, 0.83893 and 0.78347 for enhancing tumor, whole tumor and tumor core, respectively. Compared with the performance of the training and validation sets, the scores are significantly reduced. However, the high median values show that the testing set may contains some difficult cases, resulting in the lower average scores.
4 Discussion and Conclusion
We propose a S3D-UNet architecture for automatic brain tumor segmentation. In order to make full use of 3D volume information while reducing the amount of calculation, we adopt separable 3D convolutions. For the characteristics of the isotropic resolution of brain tumor MR images, we design a new separable 3D convolution architecture by dividing each 3D convolution into three branches in a parallel fashion, each with a different orthogonal view, namely axial, sagittal and coronal. We also propose a separable 3D block that takes advantage of the state-of-the-art residual inception architecture. Finally, based on separable 3D convolutions, we propose the S3D-UNet architecture using the prevalent U-Net structure.
This network has been evaluated on the BraTS 2018 Challenge testing dataset and achieved an average Dice scores of 0. 68946, 0. 83893 and 0. 78347 for the segmentation of enhancing tumor, whole tumor and tumor core, respectively. Compared with the performance of the training and validation sets, the scores of testing set are lower. This may be due to the difficult cases in testing set because the median values are high. In the future, we will work to enhance the robustness of the network.
For volumetric medical image segmentation, 3D contextual information is an important factor to obtain high-performance results. The straightforward way to capture such 3D context is to use 3D convolutions. However, the use of a large number of 3D convolutions will significantly increase the number of parameters, thus complicating the training process. In the video understanding tasks, the separable 3D convolutions with higher computational efficiency have been adopted. In this paper, we demonstrate that the U-Net with separable 3D convolutions can achieve promising results in the field of medical image segmentation.
In the future work, we will continue to improve the structure of the network and use some post-processing methods such as fully connected conditional random field to further improve the segmentation performance.
References
Menze, B.H., et al.: The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging 34, 1993–2024 (2015)
Gordillo, N., Montseny, E., Sobrevilla, P.: State of the art survey on MRI brain tumor segmentation. Magn. Reson. Imaging 31, 1426–1438 (2013)
Havaei, M., et al.: Brain tumor segmentation with Deep Neural Networks. Med. Image Anal. 35, 18–31 (2017)
Kamnitsas, K., et al.: Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 36, 61–78 (2017)
Isensee, F., Kickingereder, P., Wick, W., Bendszus, M., Maier-Hein, K.H.: Brain tumor segmentation and radiomics survival prediction: contribution to the BRATS 2017 challenge. In: Crimi, A., Bakas, S., Kuijf, H., Menze, B., Reyes, M. (eds.) BrainLes 2017. LNCS, vol. 10670, pp. 287–297. Springer, Cham (2018). https://doi.org/10.1007/978-3-319-75238-9_25
Kamnitsas, K., et al.: Ensembles of multiple models and architectures for robust brain tumour segmentation. In: Crimi, A., Bakas, S., Kuijf, H., Menze, B., Reyes, M. (eds.) BrainLes 2017. LNCS, vol. 10670, pp. 450–462. Springer, Cham (2018). https://doi.org/10.1007/978-3-319-75238-9_38
Xie, S., Sun, C., Huang, J., Tu, Z., Murphy, K.: Rethinking spatiotemporal feature learning for video understanding. arXiv:1712.04851 [cs] (2017)
Bakas, S., Reyes, M., et al.: Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge, arXiv preprint arXiv:1811.02629 (2018)
Bakas, S., et al.: Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci. Data. 4, 170117 (2017)
Bakas, S., et al.: Segmentation labels and radiomic features for the pre-operative scans of the TCGA-GBM collection. The Cancer Imaging Archive (2017)
Bakas, S., et al.: Segmentation labels and radiomic features for the pre-operative scans of the TCGA-LGG collection. The Cancer Imaging Archive (2017)
Tustison, N.J., et al.: N4ITK: improved N3 bias correction. IEEE Trans. Med. Imaging 29, 1310–1320 (2010)
Nyul, L.G., Udupa, J.K., Zhang, X.: New variants of a method of MRI scale standardization. IEEE Trans. Med. Imaging 19, 143–150 (2000)
Wang, G., Li, W., Ourselin, S., Vercauteren, T.: Automatic brain tumor segmentation using cascaded anisotropic convolutional neural networks. In: Crimi, A., Bakas, S., Kuijf, H., Menze, B., Reyes, M. (eds.) BrainLes 2017. LNCS, vol. 10670, pp. 178–190. Springer, Cham (2018). https://doi.org/10.1007/978-3-319-75238-9_16
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Çiçek, Ö., Abdulkadir, A., Lienkamp, S.S., Brox, T., Ronneberger, O.: 3D U-Net: learning dense volumetric segmentation from sparse annotation. arXiv:1606.06650 [cs] (2016)
Ulyanov, D., Vedaldi, A., Lempitsky, V.: Instance normalization: the missing ingredient for fast stylization. arXiv:1607.08022 [cs] (2016)
Sudre, C.H., Li, W., Vercauteren, T., Ourselin, S., Cardoso, M.J.: Generalised Dice overlap as a deep learning loss function for highly unbalanced segmentations. arXiv:1707.03237 [cs], vol. 10553, pp. 240–248 (2017)
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980 (2014)
Acknowledgment
This work was supported by the Department of Science and Technology of Shandong Province (Grant No. 2015ZDXX0801A01, ZR2014HQ054, 2017CXGC1502), National Natural Science Foundation of China (grant no. 61603218).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Chen, W., Liu, B., Peng, S., Sun, J., Qiao, X. (2019). S3D-UNet: Separable 3D U-Net for Brain Tumor Segmentation. In: Crimi, A., Bakas, S., Kuijf, H., Keyvan, F., Reyes, M., van Walsum, T. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2018. Lecture Notes in Computer Science(), vol 11384. Springer, Cham. https://doi.org/10.1007/978-3-030-11726-9_32
Download citation
DOI: https://doi.org/10.1007/978-3-030-11726-9_32
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-11725-2
Online ISBN: 978-3-030-11726-9
eBook Packages: Computer ScienceComputer Science (R0)