Abstract
Convolutional Neural Networks (CNN) have been obtaining successful results in the task of image segmentation in recent years. These methods use as input the sampling obtained using square uniform patches centered on each voxel of the image, which could not be the optimal approach since there is a very limited use of global context. In this work we present a new construction method for the patches by means of a circular non-uniform sampling of the neighborhood of the voxels. This allows a greater global context with a radial extension with respect to the central voxel. This approach was applied on the 2015 Longitudinal MS Lesion Segmentation Challenge dataset, obtaining better results than approaches using square uniform and non-uniform patches with the same computational cost of the CNN models.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
- Convolutional Neural Networks
- Image segmentation
- Multiple sclerosis lesions
- Magnetic resonance imaging
- Non-uniform patch
1 Introduction
Magnetic resonance imaging (MRI) of the brain has become an important tool in the medical practice due to its favourable features like the use of non-ionizant radiation, high spatial resolution, high contrast of soft tissues and the possibility to get different channels of acquisition of the same tissue. The segmentation of different tissues and structures of the brain is very useful for clinical analysis due to the fact that it allows a quantitative measurement of features present in certain diseases [6]. There is a special interest in the clinical practice of Multiple Sclerosis (MS) lesions segmentation because it allows to assist the disease diagnosis and to monitor the number and volume of the lesions [4]. The automatization of this task decreases the segmentation time considerably and could solve in part the intra and inter expert variability present in the segmentation provided by different medical practitioners.
A variety of methods have been applied to MS lesion segmentation on 3D MR images. In the last years, CNN have obtained state-of-the-art results in most of visual recognition tasks; in particular in image classification and object detection, where the input corresponds to the full image. The usual way to extend the image classification process using CNNs for a segmentation problem is using as input a patch surrounding the interest voxel to classify in one of the defined classes. The patches correspond to an intensity sampling of the neighbourhood around each voxel of the image by means of square uniform patches, where the central coordinate samples the intensity of the voxel of interest. A \(33~\times ~33\) square uniform patch is illustrated in Fig. 1(a). The size of the patch is an important parameter in the image segmentation task. On one hand, a very small patch only captures local spatial dependencies between the central voxel and the surrounding voxels, and on the other hand, the larger the patch size, the larger the context that is caught by the patch and the global spatial dependency that is taken into account at the moment of classifying the central voxel. It is important to note that increasing the patch size entails a greater computational cost, which not always ensures a considerable increment on the segmentation quality. The use of a square uniform patch implies the implicit assumption that there is not redundant information specially in the furthest voxels from the central voxel.
In the last years several methods based on CNNs for the task to MS lesion segmentation on MR images with square uniform patches [1, 2, 9] have been proposed. In [10] a cubic uniform patch was proposed as inputs. In [5] a non-uniform patch with a greater global context as input to a CNN was proposed, which resulted in an approach that outperforms the results obtained using square uniform patch as input in the same CNN architecture. The methodology for obtaining these patches is by multiplying the vectors corresponding to the coordinates of the patch by a exponential function which has its minimum point equal to 1 positioned in the center of the patch. Although this non-uniform way of expanding the patches obtain better results, the sampling of the peripheral coordinates is done with a marked asymmetry in the coordinates close to the corners, which entails that in those very distant sampling points from the central voxel, there is less significant contribution to the contextual information, since there is less spatial dependence between the voxel of interest and the more distant points. The aforementioned effect can be seen in Fig. 1(b). In this work, a new type of CNN input patches is presented, which performs a circular non-uniform sampling, that is, radially to the central voxel coordinate. This patch has two parameters to adjust, the radius of extension of the sampling and the sampling density coefficient of the local neighborhood to the coordinate of the central voxel.
(a) Square uniform patch and (b) Non-uniform patch proposed in [5].
The paper is organized as follows: In the Sect. 2 we present the proposed circular non-uniform patch, then in Sect. 3 the methodology details are discussed. In Sect. 4 we show comparative results on the 2015 Longitudinal MS Lesion Segmentation Challenge dataset. Finally, we present some concluding remarks and discuss future work.
2 Circular Non-uniform Patch
In [5] the authors propose to sample a non-uniform patch around an interest voxel, by performing a multiplication of its coordinates by an exponential function of its euclidean distance (isotropic transformation). We can see this as a transformation \(T_{NU}(\cdot )\) in Eq. (1), where the subindex NU corresponds to Non-Uniform,
where P correspond to a point in the square uniform patch Fig. 1(a), with \(P_r\) and \(P_c\) coordinates, \(\Vert \cdot \Vert _2\) is the 2-norm or euclidean distance and the only setting parameter correspond to the expansion parameter \(\gamma \). This approach applies an isotropic transformation to the sampling points with respect to the central coordinate, which leads to the fact that the sampling coordinates obtained in the furthest places or corners are greatly expanded. This implies that the sampling points of the periphery capture information with different resolution according to the angle with respect to the central voxel of the patch, which is the point of interest to classify.
In this work we propose a circular non-uniform sampling patch, which applies an anisotropic transformation to the points of the square uniform patch, generating a patch capable of extracting context in an isotropic way around the central voxel of interest. The transformation \( T_ {CNU}(\cdot )\) of the coordinates is expressed in Eq. (2), where the subindex CNU corresponds to Circular Non-Uniform,
where \(\theta _P\) is the angle of the point P. \(D(\cdot )\) is a function that returns the distance of the point belonging to the contour of the square uniform patch with the same angle, r is the parameter corresponding to the radius of the circular patch and \(\alpha \) is the parameter that determines the sampling density of the points near the center. In this way, when the point to expand corresponds to a point in the contour of the square uniform patch we will have \(\left( \frac{\Vert P\Vert _2}{D(\theta _P)}\right) ^\alpha =1\) and \(\left\| {P_r \brack P_c}\cdot \frac{r}{D(\theta _P)}\right\| _2=r\). It can be noticed that the rest of the interior points of the square uniform patch will be multiplied by values belonging to the interval \(\left[ 1,\frac{r}{D(\theta _P)}\right) \) increasing according to a power function of the parameter \(\alpha \). In Fig. 2 two patches with different values of its two parameters, radius and density coefficient are depicted.
3 Methods
The proposed and the state-of-the-art approach were evaluated on the data set of the 2015 Longitudinal MS Lesion Segmentation Challenge [2]. The data set consist of longitudinal multi-channel MR images from 19 different patients, where the training set is conformed by the MR images from five patients (segmented data set) and the test set from fourteen patients. Since in the testing set we do not have the segmentation done by experts we used only the training data set. 4.4 average time points were acquired by patient (4, 5 or 6 time points by patient) in the training and testing data sets on a 3.0 Tesla MRI scanner, where four channels were available for each patient, corresponding to T1-weighted, T2-weighted, PD-weighted and FLAIR. Each MR image has a 1[mm] cubic voxel resolution. The training data includes a binary mask with manual lesion segmentation performed by an expert.
3.1 Preprocessing
The preprocessing procedure is the previous step applied to each multi-channel MR images of the patients before to the extraction of the patch data set used as input to the CNN in the training. We can identify two preprocessing stages applied to each multi-channel MR images of the patients. The first stage corresponds to the preprocessing made by the organization on the available data set of the 2015 Longitudinal MS Lesion Segmentation Challenge [2]. This process had the following steps: (1a) Inhomogeneity bias correction, (1b) skull-stripping, (1c) dura mater stripping, (1d) a second inhomogeneity correction and (1e) rigid registration to a 1[mm] isotropic MNI template. The second stage corresponds to the following steps similar to the preprocessing made in [1]: (2a) Intensity normalization by histogram matching each MR image to a single reference case, which corresponded to the first FLAIR MR channel of the patient 1, (2b) the intensity was truncated to the percentiles in the range [0.01, 0.99], (2c) the intensity values were scaled to [0, 1], (2d) register the FLAIR image to the ICBM452 probabilistic atlas [8], (2e) the last step correspond to candidate extraction of a subset of image voxels which is performed by intersecting the sets obtained by exceeding two thresholds: (1) the intensity of FLAIR image because of the MS lesions appear as hyper-intense regions in FLAIR images [4], and (2) the probability that it belongs to the white matter due to the MS lesions are mostly located in the white matter [7].
3.2 CNN Architecture
Each convolutional block includes a convolutional layer with a specific number and size of filters and a max pooling layer of \(2\times 2\). All convolutional and fully connected layers use a Leaky ReLU activation function with a negative slope coefficient \(\beta =0.3\).
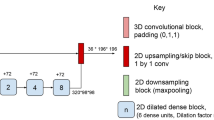
The architecture used is called Single-View Multi-Channel (SVMC), which is presented in Fig. 3. This architecture has an input layer that receives four patches of \(33\times 33\) coming from the axial view, where the patches correspond to T1-weighted, T2-weighted, PD-weighted and FLAIR channels. This patch size was chosen, due to the fact that it was the size that obtained the best results in [1]. After the third convolutional block there is a fully connected layer of sixteen nodes and as output layer a fully connected layer of two nodes with softmax activation function, which yield two output probabilities for non-lesion and lesion classes by the center voxel of the patches. This architecture is based on the structure of the V-Net CNN presented in [1].
3.3 Evaluation Metrics
To compare our approach with the state-of-the-art, we use three metrics: the True Positive Rate (TPR), Positive Predictive Value (PPV) and Dice Similarity Coefficient (Dice). TPR indicates the rate of voxels correctly segmented as lesions (Eq. (3)), PPV indicates the rate of segmented voxels from ones estimated as lesions (Eq. 4) and the Dice similarity coefficient (or F1-score, Eq. (5)) which is commonly used metric for imbalanced binary classification problems. This is an overlap measure between two binary label masks which value are in the range [0, 1], where 0 indicates no agreement and 1 means that the two masks are identical. The \(\mathcal {M_R}\) mask correspond to the lesion segmentation performed by the human rater and \(\mathcal {M_A}\) is the generated mask by the algorithm, TP, FN and FP correspond to the amount of true positive values, false negative values and false positive values.
3.4 Training Details
The weights were updated with the stochastic gradient descent and the loss function used was the categorical cross entropy function. As regularization method we used dropout with probability of 0.25 after each layer. The data set for the training was labeled into two classes, non-lesion tissue and lesion tissue. Since the voxels belonging to the non-lesion tissue are larger in quantity, it is a imbalanced classification problem. To handle this, we generated augmented data from the lesion class which balanced the data set and obtained even better results than the approach of down-sampling the majority class. The augmentation process consisted mainly in creating rotated images from angles drawn from a N(0, 5). To compare the segmentation performance of our approach and the state-of-the-art approaches, we implemented a cross patient validation where five identical models were trained, corresponding to the 5 available patients in the dataset. So, each model was trained with patches from 4 patients and was tested on the remaining 5th patient. In the training process, 80% of the patches were used to estimate the parameters and the remaining 20% were used for validation purposes (to avoid overfitting). The number of patches used for training and validation for the 5 models are: 422654, 289610, 517798, 561768 and 539002.
The implementation and trainning of the CNN was carried out using Keras Deep Neural Network Library [3] with TensorFlow as backend numerical engine. The model was trained using a NVIDIA GeForce GTX 1080TI Graphic Card.
4 Results
The cross-validation results are shown in Table 1, which corresponds to the average and standard deviation of the three metrics considered after ten cross-validations experiments on the labeled dataset, where each cross-validation experiment took around five hours. The three methods implemented and evaluated correspond to the square uniform patch (UP), the current state-of-the-art corresponding to the non-uniform patch (NUP) [5], and the proposed circular non-uniform patch (CNUP). It can be observed that the NUP with \(\gamma =0.04\) obtained the best results in the TPR metric, instead the proposed CNUP obtained the best results in PPV metric by \(r=30\) and \(\alpha =2.4\) and also by the combined Dice metric with \(r=30\) and \(\alpha =2.6\), outperforms the current state-of-the-art method corresponding to the NUP method with parameter \(\gamma =0.02\), which is the same obtained in [5]. It is important to emphasize that the Dice metric is the most used metric for the comparison of image segmentation methods.
5 Conclusions
This work shows that the quality of the context information captured by a patch depends on the manner in which the sampling of the intensity of the voxels is performed around the voxel of interest; where a non-uniform sampling with a higher sampling density near the center and with a global context sampling radially or circularly in the periphery, produces better results than current state-of-the-art proposals while preserving the same computational cost. These results can be attributable to the fact that the circular patch samples in a radial way the neighborhood of the voxel of interest, which is more similar to the spherical/ovoid shape of the lesions and the capacity of this sampling patch allows to set its extension and the density of sampling.
As future work, we will propose the possibility of extending this approach of non-uniform circular patches to non-uniform spherical patches and will analyze the option of using the temporary information present in the data set used in this work. Also we will explore the use of this approach to other medical and Synthetic Aperture Radar (SAR) images.
References
Birenbaum, A., Greenspan, H.: Multi-view longitudinal CNN for multiple sclerosis lesion segmentation. Eng. Appl. Artif. Intell. 65, 111–118 (2017)
Carass, A., et al.: Longitudinal multiple sclerosis lesion segmentation: resource and challenge. NeuroImage 148, 77–102 (2017)
Chollet, F., et al.: Keras (2015). https://keras.io
García-Lorenzo, D., Francis, S., Narayanan, S., Arnold, D.L., Collins, D.L.: Review of automatic segmentation methods of multiple sclerosis white matter lesions on conventional magnetic resonance imaging. Med. Image Anal. 17(1), 1–18 (2013)
Ghafoorian, M., et al.: Non-uniform patch sampling with deep convolutional neural networks for white matter hyperintensity segmentation. In: 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), pp. 1414–1417, April 2016
González-Villà, S., Oliver, A., Valverde, S., Wang, L., Zwiggelaar, R., Lladó, X.: A review on brain structures segmentation in magnetic resonance imaging. Artif. Intell. Med. 73, 45–69 (2016)
Inglese, M., Oesingmann, N., Casaccia, P., Fleysher, L.: Progressive multiple sclerosis and gray matter pathology: an MRI perspective. Mt. Sinai J. Med.: J. Transl. Personalized Med. 78(2), 258–267 (2011)
Mazziotta, J., et al.: A probabilistic atlas and reference system for the human brain: International consortium for brain mapping (ICBM). Philos. Trans. Royal Soc. London B: Biol. Sci. 356(1412), 1293–1322 (2001)
Roy, S., Butman, J.A., Reich, D.S., Calabresi, P.A., Pham, D.L.: Multiple sclerosis lesion segmentation from brain MRI via fully convolutional neural networks. CoRR abs/1803.09172 (2018)
Valverde, S., et al.: Improving automated multiple sclerosis lesion segmentation with a cascaded 3D convolutional neural network approach. NeuroImage 155, 159–168 (2017)
Acknowledgments
This work was supported by the Fondecyt Grant 1170123 and in part by Fondecyt Grant FB0821. Héctor Allende-Cid is supported by project Fondecyt Initiation into Research 11150248. Steren Chabert is supported by Centro de Investigación y Desarrollo en Ingeniería en Salud (CINGS).
The research of Gustavo Ulloa is also supported by the Incentive program for scientific initiation (PIIC) of the Universidad Técnica Federico Santa María DGIIP.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Ulloa, G., Naranjo, R., Allende-Cid, H., Chabert, S., Allende, H. (2019). Circular Non-uniform Sampling Patch Inputs for CNN Applied to Multiple Sclerosis Lesion Segmentation. In: Vera-Rodriguez, R., Fierrez, J., Morales, A. (eds) Progress in Pattern Recognition, Image Analysis, Computer Vision, and Applications. CIARP 2018. Lecture Notes in Computer Science(), vol 11401. Springer, Cham. https://doi.org/10.1007/978-3-030-13469-3_78
Download citation
DOI: https://doi.org/10.1007/978-3-030-13469-3_78
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-13468-6
Online ISBN: 978-3-030-13469-3
eBook Packages: Computer ScienceComputer Science (R0)