Abstract
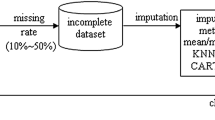
Missing Data (MD) is a common drawback when applying Data Mining on breast cancer datasets since it affects the ability of the Data mining classifier. This study evaluates the influence of MD on three classifiers: Decision tree C4.5, Support vector machine (SVM), and Multi-Layer Perceptron (MLP). For this purpose, 162 experiments were conducted using KNN imputation with three missingness mechanisms (MCAR, MAR and NMAR), and nine percentages (form 10% to 90%) applied on two Wisconsin breast cancer datasets. The MD percentage affects negatively the classifier performance. MLP achieved the lowest accuracy rates regardless the MD mechanism/percentage.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Oskouei, R.J., Kor, N.M., Maleki, S.A.: Data mining and medical world: breast cancers’ diagnosis, treatment, prognosis and challenges. Am. J. Cancer Res. (2017)
Akay, M.F.: Support vector machines combined with feature selection for breast cancer diagnosis. Expert Syst. Appl. 36, 3240–3247 (2009). https://doi.org/10.1016/j.eswa.2008.01.009
Esfandiari, N., Babavalian, M.R., Moghadam, A.M.E., Tabar, V.K.: Knowledge discovery in medicine: current issue and future trend. Expert Syst. Appl. (2014). https://doi.org/10.1016/j.eswa.2014.01.011
Idri, A., Chlioui, I., Ouassif, B.E.: A systematic map of data analytics in breast cancer. In: ACSW 2018 Proceedings pf Australasian Computer Science Week Multiconference, Brisband, pp. 26:1–26:10 (2018). https://doi.org/10.1145/3167918.3167930
Cismondi, F., Fialho, A.S., Vieira, S.M., Reti, S.R., Sousa, J.M.C., Finkelstein, S.N.: Missing data in medical databases: impute, delete or classify? Artif. Intell. Med. (2013). https://doi.org/10.1016/j.artmed.2013.01.003
Idri, A., Benhar, H., Fernández-Alemán, J.L., Kadi, I.: A systematic map of medical data preprocessing in knowledge discovery. Comput. Methods Programs Biomed. 162, 69–85 (2018). https://doi.org/10.1016/j.cmpb.2018.05.007
Idri, A., Abnane, I., Abran, A.: Missing data techniques in analogy-based software development effort estimation. J. Syst. Softw. 117, 595–611 (2016). https://doi.org/10.1016/j.jss.2016.04.058
Rubin, D.B.: Inference and missing data (with discussion). Biometrika 63, 581–592 (1976)
Garciarena, U., Santana, R.: An extensive analysis of the interaction between missing data types, imputation methods, and supervised classifiers. Expert Syst. Appl. 89, 52–65 (2017). https://doi.org/10.1016/j.eswa.2017.07.026
Curley, C., Krause, R.M., Feiock, R., Hawkins, C.V.: Dealing with missing data : a comparative exploration of approaches using the integrated city sustainability database (2017). https://doi.org/10.1177/1078087417726394
Schafer, J.L., Graham, J.W.: Missing data: our view of the state of the art. Psychol. Methods 7, 147–177 (2002). https://doi.org/10.1037/1082-989X.7.2.147
Yenduri, S.: An empirical study of imputation techniques for software data sets (2005)
García-Laencina, P.J., Abreu, P.H., Abreu, M.H., Afonoso, N.: Missing data imputation on the 5-year survival prediction of breast cancer patients with unknown discrete values. Comput. Biol. Med. 59, 125–133 (2015). https://doi.org/10.1016/j.compbiomed.2015.02.006
Jerez, J.M., Molina, I., García-Laencina, P.J., Alba, E., Ribelles, N., Martín, M., Franco, L.: Missing data imputation using statistical and machine learning methods in a real breast cancer problem. Artif. Intell. Med. 50, 105–115 (2010). https://doi.org/10.1016/j.artmed.2010.05.002
Index of /ml/machine-learning-databases/breast-cancer-Wisconsin (2017). Archive.ics.uci.edu. https://ww.archive.ics.uci.edu/ml/datasets/breast+cancer+wisconsin+(original). Accessed 20 Jul 2003
Index of /ml/machine-learning-databases/breast-cancer-wisconsin (2017). Archive.ics.uci.edu. https://archive.ics.uci.edu/ml/datasets/breast+cancer+wisconsin+(Prognostic). Accessed 20 Jul 2003
Song, Q., Shepperd, M., Chen, X., Liu, J.: Can k-NN imputation improve the performance of C4.5 with small software project data sets? a comparative evaluation. J. Syst. Softw. (2008). https://doi.org/10.1016/j.jss.2008.05.008
Hall, M., Witten, I., Frank, E.: Data Mining, 4th Edn., Elsevier (2011)
Alpaydın, E.: Introduction to Machine Learning, 2nd Edn., The MIT Press, London (2014). https://doi.org/10.1007/978-1-62703-748-8-7
Cristianini, N., Shawe-Taylor, J.: An Introduction to Support Vector Machines and other kernel based learning methods. Cambridge University Press, Cambridge (2000). citeulike-article-id:114719
Ghosh, S., Mondal, S., Ghosh, B.: A comparative study of breast cancer detection based on SVM and MLP BPN classifier. In: 2014 First International Conference on Automation, Control, Energy and System, pp. 1–4 (2014). https://doi.org/10.1109/aces.2014.6808002
Baldi, P., Brunak, S., Chauvin, Y., Andersen, C.A., Nielsen, H.: Assessing the accuracy of prediction algorithms for classification: an overview. Bioinformatics (2000). https://doi.org/10.1093/bioinformatics/16.5.412
Fawcett, T.: An introduction to ROC analysis. Pattern Recognit. Lett. (2006). https://doi.org/10.1016/j.patrec.2005.10.010
Salzberg, S.L.: On comparing classifiers: Pitfalls to avoid and a recommended approach. Data Min. Knowl. Discov. (1997). https://doi.org/10.1023/a:1009752403260
Jhajharia, S., Varshney, H.K., Verma, S., Kumar, R.: A neural network based breast cancer prognosis model with PCA processed features. In: 2016 International Conference on Advances in Computing, Communications and Informatics, ICACCI 2016, pp. 1896–1901 (2016). https://doi.org/10.1109/ICACCI.2016.7732327
The university of Waikato, Weka the university of Waikato, (n.d.). https://www.cs.waikato.ac.nz/ml/weka/
Ma, X., Zhang, Y., Wang, Y.: Performance evaluation of kernel functions based on grid search for support vector regression. In: 2015 IEEE 7th International Conference on Cybernetics and Intelligent Systems and IEEE Conference on Robotics, Automation and Mechatronics, pp. 283–288 (2015). https://doi.org/10.1109/ICCIS.2015.7274635
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Chlioui, I., Idri, A., Abnane, I., de Gea, J.M.C., Fernández-Alemán, J.L. (2019). Breast Cancer Classification with Missing Data Imputation. In: Rocha, Á., Adeli, H., Reis, L., Costanzo, S. (eds) New Knowledge in Information Systems and Technologies. WorldCIST'19 2019. Advances in Intelligent Systems and Computing, vol 932. Springer, Cham. https://doi.org/10.1007/978-3-030-16187-3_2
Download citation
DOI: https://doi.org/10.1007/978-3-030-16187-3_2
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-16186-6
Online ISBN: 978-3-030-16187-3
eBook Packages: Intelligent Technologies and RoboticsIntelligent Technologies and Robotics (R0)