Abstract
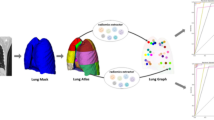
Pulmonary tuberculosis (TB) is still an important cause of death worldwide, even after being almost eradicated 40 years ago. Early identification of TB in computed tomography (CT) scans can influence therapeutical decisions, thus improving patient outcome. In this paper, a graph model of the lungs is proposed for the classification of TB types using local texture features in thorax CT scans of TB patients. Based on lung morphology, an automatic patient-specific lung field parcellation was initially computed. Local visual features were then extracted from each region and were used to build a personalized lung graph model. A new graph-based patient descriptor enables comparisons between lung graphs with a different number of nodes and edges, encoding the distribution of several node measures from graph theory. The proposed model was trained and tested on a public dataset of 1,513 CT scans of 994 TB patients. The evaluation was performed on data from a scientific challenge together with 39 participant algorithms, obtaining the best unweighted Cohen kappa coefficient of 0.24 in a 5-class setup with 505 test CTs. Even though each lung graph has a unique structure, the proposed method was able to identify key texture changes associated with the different manifestations of TB.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Notes
- 1.
https://www.imageclef.org/2017/tuberculosis, as of 1 March 2019.
References
Achanta, R., Shaji, A., Smith, K., Lucchi, A., Fua, P., Süsstrunk, S.: SLIC superpixels compared to state-of-the-art superpixel methods. IEEE Trans. Pattern Anal. Mach. Intell. 34(11), 2274–2282 (2012)
Andreu, J., Caceres, J., Pallisa, E., Martinez-Rodriguez, M.: Radiological manifestations of pulmonary tuberculosis. Eur. J. Radiol. 51(2), 139–149 (2004)
Blumberg, H.M., Burman, W.J., Chaisson, R.E., Daley, C.L., et al.: American thoracic society/centers for disease control and prevention/infectious diseases society of America: treatment of tuberculosis. Am. J. Respir. Crit. Care Med. 167(4), 603 (2003)
Burrill, J., Williams, C.J., Bain, G., Conder, G., Hine, A.L., Misra, R.R.: Tuberculosis: a radiologic review. Radiographics 27(5), 1255–1273 (2007)
Dicente Cid, Y., Batmanghelich, K., Müller, H.: Textured graph-based model of the lungs: application on tuberculosis type classification and multi-drug resistance detection. In: Bellot, P., et al. (eds.) CLEF 2018. LNCS, vol. 11018, pp. 157–168. Springer, Cham (2018). https://doi.org/10.1007/978-3-319-98932-7_15
Dicente Cid, Y., Jimenez-del-Toro, O., Depeursinge, A., Müller, H.: Efficient and fully automatic segmentation of the lungs in CT volumes. In: Goksel, O., Jimenez-del-Toro, O., Foncubierta-Rodriguez, A., Müller, H. (eds.) Proceedings of the VISCERAL Challenge at ISBI, pp. 31–35. No. 1390 in CEUR Workshop Proceedings, April 2015
Dicente Cid, Y., Jiménez-del-Toro, O., Platon, A., Müller, H., Poletti, P.-A.: From local to global: a holistic lung graph model. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11071, pp. 786–793. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00934-2_87
Dicente Cid, Y., Kalinovsky, A., Liauchuk, V., Kovalev, V., Müller, H.: Overview of ImageCLEFtuberculosis 2017 - predicting tuberculosis type and drug resistances. In: CLEF 2017 Labs Working Notes. CEUR Workshop Proceedings, 11–14 September 2017, Dublin, Ireland. CEUR-WS.org (2017). http://ceur-ws.org
Dicente Cid, Y., Liauchuk, V., Kovalev, V., Müller, H.: Overview of image CLEF tuberculosis 2018 - detecting multi-drug resistance, classifying tuberculosis type, and assessing severity score. In: CLEF 2018 Working Notes. CEUR Workshop Proceedings, 10–14 September 2018, Avignon, France. CEUR-WS.org (2018). http://ceur-ws.org
Dicente Cid, Y., Müller, H.: Texture-based graph model of the lungs for drug resistance detection, tuberculosis type classification, and severity scoring: Participation in Image CLEF 2018 tuberculosis task. In: CLEF 2018 Working Notes. CEUR Workshop Proceedings, 10–14 September 2018, Avignon, France. CEUR-WS.org (2018). http://ceur-ws.org
Dicente Cid, Y., Müller, H., Platon, A., Poletti, P.A., Depeursinge, A.: 3-D solid texture classification using locally-oriented wavelet transforms. IEEE Trans. Image Process. 26(4), 1899–1910 (2017)
Ishay, A., Marques, O.: Ensemble of 3D CNNs with multiple inputs for tuberculosis type classification. In: CLEF 2018 Working Notes. CEUR Workshop Proceedings, 10–14 September 2018, Avignon, France. CEUR-WS.org (2018). http://ceur-ws.org
Liauchuk, V., Tarasau, A., Snezhko, E., Kovalev, V., Gabrielian, A., Rosenthal, A.: ImageCLEF 2018: Lesion-based TB-descriptor for CT image analysis. In: CLEF2018 Working Notes. CEUR Workshop Proceedings, 10–14 September 2018, Avignon, France. CEUR-WS.org (2018). http://ceur-ws.org
Liu, K., Skibbe, H., Schmidt, T., Blein, T., Palme, K., Brox, T., Ronneberger, O.: Rotation-invariant hog descriptors using fourier analysis in polar and spherical coordinates. Int. J. Comput. Vis. 106(3), 342–364 (2014)
Parra, J.A.C., Zúñiga, N.M., Lara, C.S.: Tuberculosis “the great imitator”: false healing and subclinical activity. Indian J. Tuberc. 64(4), 345–348 (2017)
Richiardi, J., Bunke, H., Van De Ville, D., Achard, S.: Machine learning with brain graphs. IEEE Sig. Process. Mag. 30, 58 (2013)
Richiardi, J., Eryilmaz, H., Schwartz, S., Vuilleumier, P., Van De Ville, D.: Decoding brain states from fMRI connectivity graphs. NeuroImage 56(2), 616–626 (2011). https://doi.org/10.1016/j.neuroimage.2010.05.081
Schabdach, J., Wells, W.M., Cho, M., Batmanghelich, K.N.: A likelihood-free approach for characterizing heterogeneous diseases in large-scale studies. In: Niethammer, M., et al. (eds.) IPMI 2017. LNCS, vol. 10265, pp. 170–183. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-59050-9_14
Sun, J., Chong, P., Tan, Y.X.M., Binder, A.: ImageCLEF 2017: ImageCLEF tuberculosis task - the SGEast submission. In: CLEF 2017 Working Notes. CEUR Workshop Proceedings, 11–14 September 2017, Dublin, Ireland. CEUR-WS.org (2017). http://ceur-ws.org
Varoquaux, G., Gramfort, A., Poline, J., Thirion, B.: Brain covariance selection: better individual functional connectivity models using population prior. In: NIPS, vol. 10, pp. 2334–2342 (2010)
World Health Organization, et al.: Global tuberculosis report 2016 (2016)
Acknowledgments
This work was partly supported by the Swiss National Science Foundation in the PH4D project (grant agreement 320030-146804).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Dicente Cid, Y., Jimenez-del-Toro, O., Poletti, PA., Müller, H. (2019). A Graph Model of the Lungs with Morphology-Based Structure for Tuberculosis Type Classification. In: Chung, A., Gee, J., Yushkevich, P., Bao, S. (eds) Information Processing in Medical Imaging. IPMI 2019. Lecture Notes in Computer Science(), vol 11492. Springer, Cham. https://doi.org/10.1007/978-3-030-20351-1_28
Download citation
DOI: https://doi.org/10.1007/978-3-030-20351-1_28
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-20350-4
Online ISBN: 978-3-030-20351-1
eBook Packages: Computer ScienceComputer Science (R0)