Abstract
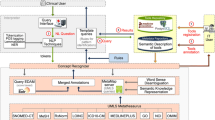
This paper proposes an ontology learning framework that combines text mining, information extraction and retrieval. The proposed model takes advantage of existing structured knowledge by reusing terms and concepts from other ontologies. We further apply the methodology to create a detailed ontology for the emerging precision medicine (PM) domain by collecting a corpus of relevant articles and mapping its frequent terms to existing concepts. The resulting ontology consists of 543 annotated classes. The ontology was also tested for effectiveness by applying two evaluation frameworks to validate its design and quality. The results demonstrate that the ontology learning system is able to capture and represent the semantics of the PM domain with high precision and significance. Moreover, the computer-assisted construction process reduced dependency on expert knowledge. The developed PreMedOnto ontology could be further used to enhance the potentials of other NLP applications in the PM domain.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Ali-Khan, S., Kowal, S., Luth, W., Gold, R., Bubela, T.: Terminology for personalized medicine: a systematic collection terminology for personalized medicine. Technical report (2016)
Alobaidi, M., Malik, K.M., Hussain, M.: Automated ontology generation framework powered by linked biomedical ontologies for disease-drug domain. Comput. Methods Programs Biomed. 165, 117–128 (2018). https://doi.org/10.1016/j.cmpb.2018.08.010
Alobaidi, M., Malik, K.M., Sabra, S.: Linked open data-based framework for automatic biomedical ontology generation. BMC Bioinform. 19(1), 319 (2018). https://doi.org/10.1186/s12859-018-2339-3
Amato, F., Santo, A.D., Moscato, V., Picariello, A., Serpico, D., Sperli, G.: A lexicon-grammar based methodology for ontology population for e-health applications. In: 2015 Ninth International Conference on Complex, Intelligent, and Software Intensive Systems. pp. 521–526. IEEE, July 2015. https://doi.org/10.1109/CISIS.2015.76
Arguello Casteleiro, M., et al.: Deep learning meets ontologies: experiments to anchor the cardiovascular disease ontology in the biomedical literature. J. Biomed. Semant. 9(1), 13 (2018). https://doi.org/10.1186/s13326-018-0181-1
Bontas, E.P., Mochol, M., Tolksdorf, R.: Case Studies on Ontology Reuse. Technical report
Buitelaar, P., Cimiano, P., Magnini, B.: Ontology learning from text: methods. Eval. Appl. (2005). https://doi.org/10.1162/coli.2006.32.4.569
Cahyani, D.E., Wasito, I.: Automatic ontology construction using text corpora and ontology design patterns (ODPs) in Alzheimer’s disease. Jurnal Ilmu Komputer dan Informasi 10(2), 59 (2017). https://doi.org/10.21609/jiki.v10i2.374
Dramé, K., et al.: Reuse of termino-ontological resources and text corpora for building a multilingual domain ontology: an application to Alzheimer’s disease. J. Biomed. Inform. 48, 171–182 (2014). https://doi.org/10.1016/J.JBI.2013.12.013
Duque-ramos, A., Duque-ramos, A., Fernández-breis, J.T., Stevens, R., Aussenac-gilles, N.: OQuaRE: a SQuaRE-based approach for evaluating the quality of ontologies. J. Res. Pract. Inf. Technol. 43, 159 (2011)
Gao, M., Chen, F., Wang, R.: Improving Medical Ontology Based on Word Embedding (2018). https://doi.org/10.1145/3194480.3194490
Gedzelman, S., Simonet, M., Bernhard, D., Diallo, G., Palmer, P.: Building an ontology of cardio-vascular diseases for concept-based information retrieval. In: Computers in Cardiology, 2005, pp. 255–258. IEEE (2005). https://doi.org/10.1109/CIC.2005.1588085
Gruber, T.R.: A translation approach to portable ontology specifications. Knowl. Acquisition 5(2), 199–220 (1993). https://doi.org/10.1006/KNAC.1993.1008
Jiménez-Ruiz, E., Cuenca Grau, B., Sattler, U., Schneider, T., Berlanga, R.: Safe and Economic Re-Use of Ontologies: A Logic-Based Methodology and Tool Support. Technical report
Kang, Y., Fink, J.C., Doerfler, R., Zhou, L.: Disease specific ontology of adverse events: ontology extension and adaptation for chronic kidney disease. Comput. Biol. Med. 101, 210–217 (2018). https://doi.org/10.1016/J.COMPBIOMED.2018.08.024
Lossio-Ventura, J.A., Jonquet, C., Roche, M., Teisseire, M.: A Way to Automatically Enrich Biomedical Ontologies. https://doi.org/10.5441/002/edbt.2016.82
Ochs, C., Perl, Y., Geller, J., Arabandi, S., Tudorache, T., Musen, M.A.: An empirical analysis of ontology reuse in BioPortal. J. Biomed. Inform. 71, 165–177 (2017). https://doi.org/10.1016/J.JBI.2017.05.021
Poveda-Villalón, M., Carmen Suárez-Figueroa, M., Ángel García-Delgado, M., Gómez-Pérez, A.: OOPS! (OntOlogy Pitfall Scanner!): supporting ontology evaluation on-line. Technical report (2009)
Sánchez, D., Moreno, A.: Learning medical ontologies from the Web. Technical report
Shah, T., Rabhi, F., Ray, P., Taylor, K.: A guiding framework for ontology reuse in the biomedical domain. In: 2014 47th Hawaii International Conference on System Sciences, pp. 2878–2887. IEEE January 2014. https://doi.org/10.1109/HICSS.2014.360
Xiang, Z., Courtot, M., Brinkman, R.R., Ruttenberg, A., He, Y.: OntoFox: web-based support for ontology reuse. BMC Res. Notes 3(1), 175 (2010). https://doi.org/10.1186/1756-0500-3-175
Yates, L.R., et al.: The european society for medical oncology (ESMO) precision medicine glossary. Ann. Oncol. 29(1), 30–35 (2018). https://doi.org/10.1093/annonc/mdx707
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Tawfik, N.S., Spruit, M.R. (2019). PreMedOnto: A Computer Assisted Ontology for Precision Medicine . In: Métais, E., Meziane, F., Vadera, S., Sugumaran, V., Saraee, M. (eds) Natural Language Processing and Information Systems. NLDB 2019. Lecture Notes in Computer Science(), vol 11608. Springer, Cham. https://doi.org/10.1007/978-3-030-23281-8_28
Download citation
DOI: https://doi.org/10.1007/978-3-030-23281-8_28
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-23280-1
Online ISBN: 978-3-030-23281-8
eBook Packages: Computer ScienceComputer Science (R0)