Abstract
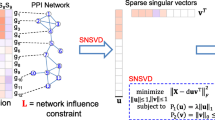
Sparse Singular Value Decomposition (SSVD) model has been proposed to bicluster gene expression data to identify gene modules. However, traditional SSVD model can only handle the gene expression data where no gene-gene interaction information is integrated. Here, we develop a Sparse Network-regularized SVD (SNSVD) method, which can integrate the gene-gene interaction information into the SSVD model, to identify the underlying gene functional modules from the gene expression data. The simulation results on synthetic data show that SNSVD is more effective than the traditional SVD-based methods.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Lee, M.S., et al.: Biclustering via sparse singular value decomposition. Biometrics 66(4), 1087–1095 (2010)
Liquet, B., et al.: Group and sparse group partial least square approaches applied in genomics context. Bioinformatics 32(1), 35–42 (2015)
Min, W., et al.: A two-stage method to identify joint modules from matched MicroRNA and mRNA expression data. IEEE Trans. Nanobiosci. 15(4), 362–370 (2016)
Eren, K., et al.: A comparative analysis of biclustering algorithms for gene expression data. Brief. Bioinform. 14(3), 279–292 (2013)
Sill, M.K., et al.: Robust biclustering by sparse singular value decomposition incorporating stability selection. Bioinformatics 27(15), 2089–2097 (2011)
Oghabian, A., et al.: Biclustering methods: biological relevance and application in gene expression analysis. PLOS One 9(3), e90801 (2014)
Chen, S., Liu, J., Zeng, T.: Measuring the quality of linear patterns in biclusters. Methods 83, 18–27 (2015)
Yang, D., Ma, Z., Buja, A.: Rate optimal denoising of simultaneously sparse and low rank matrices. J. Mach. Learn. Res. 17(1), 3163–3189 (2016)
Asteris, M.K., et al.: A simple and provable algorithm for sparse diagonal CCA. In: 33rd International Conference on Machine Learning, New York, NY, USA, pp. 1148–1157 (2016)
Chuang, H., et al.: Network-based classification of breast cancer metastasis. Mol. Syst. Biol. 3, 140 (2007)
Sokolov, A., et al.: Pathway-based genomics prediction using generalized elastic net. PLoS Comput. Biol. 12, 3 (2016)
Hill, S.M., et al.: Inferring causal molecular networks: empirical assessment through a community-based effort. Nat. Methods 13(4), 310–318 (2016)
Glaab, E.: Using prior knowledge from cellular pathways and molecular networks for diagnostic specimen classification. Brief. Bioinform. 17(3), 440–452 (2016)
Lee, E., et al.: Inferring pathway activity toward precise disease classification. PLoS Comput. Biol. 4, 11 (2008)
Li, C., et al.: Network-constrained regularization and variable selection for analysis of genomic data. Bioinformatics 24(9), 1175–1182 (2008)
Iorio, F., et al.: A landscape of pharmacogenomic interactions in cancer. Cell 166(3), 740–754 (2016)
Cerami, E.G., et al.: Pathway commons, a web resource for biological pathway data. Nucleic Acids Res. 39, D685–D690 (2011)
Bolte, J., et al.: Proximal alternating linearized minimization for nonconvex and nonsmooth problems. Math. Program. 146(1), 459–494 (2014)
Friedman, J.H., et al.: Pathwise coordinate optimization. Ann. Appl. Stat. 1(2), 302–332 (2007)
Fan, J., et al.: Variable selection via nonconcave penalized likelihood and its oracle properties. J. Am. stat. Assoc. 96(456), 1348–1360 (2001)
Huang, D., et al.: Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4(1), 44–57 (2009)
Leeksma, O.C., et al.: Germline mutations predisposing to diffuse large B-cell lymphoma. Blood Cancer J. 7(2), e532 (2017)
Disis, M.L.: Immune Regulation of Cancer. J. Clin. Oncol. 28(29), 4531–4538 (2010)
Acknowledgement
This work has been supported by the National Natural Science Foundation of China 61802157, Youth Project from the Education Department of Jiangxi Province of China GJJ180626.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Zhu, F., Liu, J., Min, W. (2019). Gene Functional Module Discovery via Integrating Gene Expression and PPI Network Data. In: Huang, DS., Jo, KH., Huang, ZK. (eds) Intelligent Computing Theories and Application. ICIC 2019. Lecture Notes in Computer Science(), vol 11644. Springer, Cham. https://doi.org/10.1007/978-3-030-26969-2_11
Download citation
DOI: https://doi.org/10.1007/978-3-030-26969-2_11
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-26968-5
Online ISBN: 978-3-030-26969-2
eBook Packages: Computer ScienceComputer Science (R0)