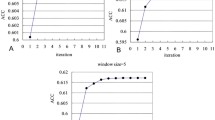
Abstract
The function of G-protein-coupled receptor (GPCR) in organisms are directly related to its tertiary structure. Protein free energy can well reflect the state stability of protein tertiary structure. Therefore, the research on the free energy of GPCR is of great significance. At present, there is a lack of goldenfree energy constraint GPCR structure in helix domain level in the current researches, which affects GPCR’s three-dimensional structure stability. In this paper, a knowledge-based free energy term with the residue distance and angle of the helix of the GPCR as a constraint is established. The energy term is based on the gaussian distribution model, which accurately expresses the free energy of the GPCR. Compared with other energy functions, the experimental data and the result of this model is more accurate.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Hauser, A.S., Attwood, M.M., Rask-Andersen, M., et al.: Trends in GPCR drug discovery: new agents, targets and indications. Nat. Rev. Drug Discov. 16, 829–842 (2017)
Flock, T., Hauser, A.S., Lund, N., et al.: Selectivity determinants of GPCR–G-protein binding. Nature 545(7654), 317–322 (2017)
Hilger, D., Masureel, M., Kobilka, B.K.: Structure and dynamics of GPCR signaling complexes. Nat. Struct. Mol. Biol. 25(1), 4–12 (2018)
Kang, Y., Zhou, X.E., Xiang, G., et al.: Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature 523(7562), 561–567 (2015)
Moritsugu, K., Terada, T., Kidera, A.: Free-energy landscape of protein-ligand interactions coupled with protein structural changes. J. Phys. Chem. B 121(4), 731–740 (2017)
Takemura, K., Matubayasi, N., Kitao, A.: Binding free energy analysis of protein-protein docking model structures by evERdock. J. Chem. Phys. 148(10), 105101 (2018)
Gohlke, H., Kiel, C., Case, D.A.: Insights into protein-protein binding by binding free energy calculation and free energy decomposition for the Ras-Raf and Ras-RalGDS complexes. J. Mol. Biol. 330(4), 891–913 (2003)
Lee, H.S., Seok, C., Im, W.: Potential application of alchemical free energy simulations to discriminate GPCR ligand efficacy. J. Chem. Theory Comput. 11(3), 1255–1266 (2015)
Lenselink, E.B., Louvel, J., Forti, A.F., et al.: Predicting binding affinities for GPCR ligands using free-energy perturbation. ACS Omega 1(2), 293–304 (2016)
Advances in free-energy-based simulations of protein folding and ligand binding. Curr. Opin. Struct. Biol. 36, 25–31 (2016)
Suofu, Y., Li, W., Jeanalphonse, F.G., et al.: Dual role of mitochondria in producing melatonin and driving GPCR signaling to block cytochrome c release. Proc. Natl. Acad. Sci. U.S.A. 114(38), E7997–E8006 (2017)
Pavlos, N.J., Friedman, P.A.: GPCR signaling and trafficking: the long and short of it. Trends Endocrinol. Metab. 28(3), 213–226 (2017)
Irannejad, R., Pessino, V., Mika, D., et al.: Functional selectivity of GPCR-directed drug action through location bias. Nat. Chem. Biol. 13(7), 799 (2017)
Eichel, K., Jullié, D., Barsirhyne, B., et al.: Catalytic activation of β-arrestin by GPCRs. Nature 557, 381–386 (2018)
Jean-Charles, P.Y., Kaur, S., Shenoy, S.K.: GPCR signaling via β-arrestin-dependent mechanisms. J. Cardiovasc. Pharmacol. 70(3), 142–158 (2017)
Kumar, B.A., Kumari, P., Sona, C., et al.: GloSensor assay for discovery of GPCR-selective ligands. Methods Cell Biol. 142, 27–50 (2017)
Pándy-Szekeres, G., Munk, C., Tsonkov, T.M., et al.: GPCRdb in 2018: adding GPCR structure models and ligands. Nucleic Acids Res. 46(Database issue), 440–446 (2017)
Mcgregor, K.M., Bécamel, C., Marin, P., et al.: Using melanopsin to study G protein signaling in cortical neurons. J. Neurophysiol. 116(3), 1082–1092 (2016)
Huang, Y., Todd, N., Thathiah, A.: The role of GPCRs in neurodegenerative diseases: avenues for therapeutic intervention. Curr. Opin. Pharmacol. 32, 96–110 (2017)
Gupta, A., Singh, V.: GPCR Signaling in C. Elegans and its implications in immune response. Adv. Immunol. 136, 203–226 (2017)
Acknowledgement
This paper is supported by the National Natural Science Foundation of China (61772357, 61502329, 61672371, and 61876217), Jiangsu Province 333 Talent Project, Top Talent Project (DZXX-010), Suzhou Foresight Research Project (SYG201704, SNG201610, and SZS201609)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Ling, H., Wu, H., Han, J., Ding, J., Lu, W., Fu, Q. (2019). Knowledge Based Helix Angle and Residue Distance Restraint Free Energy Terms of GPCRs. In: Huang, DS., Jo, KH., Huang, ZK. (eds) Intelligent Computing Theories and Application. ICIC 2019. Lecture Notes in Computer Science(), vol 11644. Springer, Cham. https://doi.org/10.1007/978-3-030-26969-2_16
Download citation
DOI: https://doi.org/10.1007/978-3-030-26969-2_16
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-26968-5
Online ISBN: 978-3-030-26969-2
eBook Packages: Computer ScienceComputer Science (R0)