Abstract
In this work, we aimed at predicting children’s fluid intelligence scores based on structural T1-weighted MR images from the largest long-term study of brain development and child health. The target variable was regressed on a data collection site, sociodemographic variables, and brain volume, thus being independent to the potentially informative factors, which were not directly related to the brain functioning. We investigated both feature extraction and deep learning approaches as well as different deep CNN architectures and their ensembles. We proposed an advanced architecture of VoxCNNs ensemble, which yields MSE (92.838) on a blind test.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
- MRI analysis
- Fluid intelligence prediction
- Deep learning
- 3D convolutional neural networks
- VoxCNN ensemble
1 Introduction
Understanding cognitive development in children may potentially improve their health outcomes through adolescence. Thus, determining neural mechanism underlying general intelligence is a critical task. One of two discrete factors of general intelligence is fluid intelligence.
Fluid intelligence is the capacity to think logically and solve problems in novel situations, independent of acquired knowledge. It involves the ability to identify patterns and relationships that underpin novel problems and to extrapolate these findings using logic [1].
There are studies on fluid intelligence prediction based on various brain imaging techniques and extracted features [23, 40]. However, the authors could not highlight robust biomarkers and methods to predict fluid intelligence scores .
Deep learning approaches and convolutional neural networks, in particular, have shown high potential on imagery classification, recognition and processing and thus could be considered useful for fluid intelligence scores prediction based on MRI data (3D brain images).
The advantage of deep learning methods is the ability to automatically derive complex and informative features from the raw data during the training process. That allows training a neural network directly on high-dimensional 3D brain imaging data skipping the feature extraction step.
By design, neural architectures for deep learning are built in a modular way, with basic building blocks, such as composite convolutional layers, typically reused across many models and applications. This enables the standardization of deep learning architectures, with much research devoted to the exploration of pre-built layers and pre-trained activations (for transfer learning, image retrieval, etc.). However, the choice of appropriate architecture targeting specific clinical applications such as cognitive potential prediction or pathology classification remains open problem and requires further investigation.
In the present study we carried out an extensive experimental evaluation of deep voxelwise neural network architectures for fluid intelligence scores prediction based on MRI data with multimodal input structure.
The article has the following structure. In Sect. 2 we review deep network architectures used for MRI data processing. In Sect. 3 we present the training dataset and our deep network architecture. We describe obtained results in Sect. 4, provide discussions in Sect. 5 and draw conclusions in Sect. 6.
2 Related Work
There is a number of successful applications of convolutional neural networks (CNN) with different architectures for segmentation of MRI data. Many of these solutions are based on adapting existing approaches to analyzing 2D images for processing of three-dimensional data.
For example, for brain segmentation, an architecture similar to ResNet [20] was proposed, which expands the possibilities of deep residual learning for processing volumetric MRI data using 3D filters in convolutional layers. The model, called VoxResNet [32], consists of volumetric residual blocks (VoxRes blocks), containing convolutional layers as well as several deconvolutional layers. The authors demonstrated the potential of ResNet-like volumetric architectures, achieving better results than many modern methods of MRI image segmentation [22]. Convolutional neural networks also showed good classification results in problems associated with neuropsychiatric diseases such as Alzheimer’s disease.
Recently proposed classification model with a VGG-like architecture called VoxCNN was used for neuro-degenerative decease classification [21]. These results were more accurate or comparable to earlier approaches that use previously extracted morphometrical lower dimensional brain characteristics [34, 38, 39].
Thus, convolutional networks can be applied directly to the raw neuroimaging data without loss of model performance and over-fitting, which allows skipping the pre-processing step.
However, to the depth of our knowledge, there has not been much work on the use of convolutional networks for predicting fluid intelligence based on MRI imaging.
3 Materials and Methods
3.1 Data Set
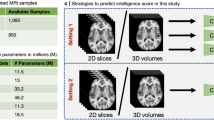
The training data set was provided by ABCD Neurocognitive Prediction Challenge (ABCD-NP-Challenge 2019Footnote 1). The dataset consists of T1-weighed MR brain images of four thousand individuals (of age 9–10 years) as well as corresponding sociodemographic variables [33]. The participants’ fluid intelligence scores (4154 subjects, 3739 for training and 415 for validation) were also provided.
3.2 Target Processing
The fluid intelligence scores were pre-residualized on a data collection site, sociodemographic variables and brain volume. For that a linear regression model was fitted with fluid intelligence as the dependent variable and brain volume, data collection site, age at baseline, sex at birth, race/ethnicity, highest parental education, parental income, and parental marital status as independent variables [33].
The obtained residuals were used as targets to be predicted by a neural network. This approach is known to be used in GML models, for fMRI data analysis, allowing removal of linear dependencies between dependent variables.
3.3 MRI Data Processing
Imagery dataset consists of skull stripped images affinely aligned to the SRI 24 atlas [5], segmented into regions of interest according to the atlas, and the corresponding volume scores of each ROI [29]. T1-weighted MRI was transformed according to the Minimal Processing Pipeline by ABCD [33].
The cross-sectional component of the National Consortium on Alcohol and NeuroDevelopment in Adolescence (NCANDA) pipeline [12] was applied to T1 images. The steps included noise removal and field inhomogeneity correction confined to the brain mask, defined by non-rigidly aligning SRI24 atlas to the T1w MRI via Advanced normalization tools (ANTS) [4].
The brain mask was refined by majority voting across maps extracted by FSL BET [3], AFNI 3dSkullStrip [2], FreeSurfer mrigcut [6], and the Robust Brain Extraction (ROBEX) methods [8], which were applied on combinations of bias and non-bias corrected T1w images. Using the refined masked, image inhomogeneity correction was repeated and the skull-stripped T1w image was segmented into brain tissue (gray matter, white matter, and cerebrospinal fluid) via Atropos [7]. Gray matter tissue was further parcelled according to the SRI24 atlas, which was non-rigidly registered to the T1w image via ANTS.
3.4 Specifications of the Investigated Models
We use an ensemble of deep neural networks with VoxCNN architecture [27, 37] to solve the regression problem. The proposed architecture has already demonstrated some successful applications to brain image analysis tasks. To provide better convergence and stronger regularization of results we enhanced this architecture.
VoxCNN networks are similar to VGG [11] architecture, which is a popular architecture for 2D-images classification. VoxCNN applies 3D convolutions to deal with three-dimensional MRI brain scans.
Proposed network consists of four blocks with two convolutional layers each having 3D convolutions followed by batch-normalization and ReLU activation function [41]. Number of filters in convolutional layers starts from 16 in the first block and doubles with each next block. Filters of the very first layer are applied with the stride x2 to reduce the dimension of the original image. Our experiments have shown that this step does not reduce the network performance but helps to speed up the convergence and meet the limitations of GPU memory. The blocks are separated by max-pooling layers. We also apply 3D-dropout after each pooling layer to promote independence between feature maps and reduce over-fitting [15].
Next, feature maps extracted by the convolutional layers are fed into the fully connected layer with 1024 hidden units, batch-normalization, ReLU activation, and dropout regularization, and then to the final layer with a single unit without non-linearity.
It was previously shown that auxiliary tower backpropagates the classification loss earlier in the network, serving as an additional regularization mechanism [14, 24].
Therefore, the auxiliary output was added to the network to provide better training of the deeper layers. For this purpose, feature maps from intermediate layers were fed to the separate fully connected layer to produce another target prediction, which was then added to the main network output with adjusted weight. In this case, the output of the third block of convolutional layer was used to compute auxiliary prediction and average it with the main output with weights 0.4 and 0.6 respectively.
We assessed model quality by Mean Squared Error (MSE) between the predicted scores and the pre-residualized fluid intelligence scores. The models were selected by optimizing the MSE-loss with the Adam optimizer. The learning rate was set to 3e-5, batch size is 10 and each network was trained until the loss on validation set starts to increase.
To train the model we used multi-modal input data: brain scan data (T1-weighted imagery after preprocessing) and gray matter segmented brain masks. For each subject, two three-dimensional images were stacked as channels of a single image. We fed the resulted 3D image with two channels into the VoxCNN network as an input.
We used cross-validation to increase the model performance: we split the training sample into two separate parts and two neural networks are trained with the same architecture on each part independently. Then for the validation subjects, an ensemble of these two models, defined as a weighted average of their predictions, was applied. Weights for averaging were determined based on the validation performance of each model (test predictions of the network that turned out to demonstrate lower MSE score on validation were set to larger weights). The number of layers, Stride and ReLU blocks position were adjusted correspondingly (Fig. 1).
The train set consists of n = 3739 samples, the validation set – n = 415 samples, and the test set – n = 4515 samples.
The models were implemented in PyTorch and trained on a single GPU [18].
4 Experimental Results
In Table 1 the explored deep neural network architectures are specified as well as corresponding results for fluid intelligence prediction. Here the brain morphometric characteristics predictive capacity is considered as a baseline for prediction.
The most accurate prediction (in terms of MSE on the validation set) was obtained as a weighted average of the two predictions by VoxCNN trained on different parts of the training sample:
-
1.
VoxCNN network, trained on both brain T1 images and segmented images,
-
2.
VoxCNN network (with auxiliary head for better convergence), trained on brain T1 images, segmented images and additional socio-demographic data. We used segmented brain masks and full brain imagery after pre-processing.
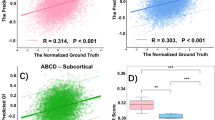
As a result, the first and the second network architectures showed 71.777 and 71.094 MSE scores on the Validation set. After averaging the predictions with adjusted weights \(\frac{2}{3}\) and \(\frac{1}{3}\), the final validation performance reached 70.635 MSE when using ensembles of models.
Then on the Test set the ensemble models yielded 92.8378 and 94.0808 MSE scores correspondingly (Table 2).
5 Discussion
All considered regression models provided MSE close to 70. These results are comparable to the baseline result, calculated using morphological characteristics on the Validation set.
This incremental improvement and rather high errors across all models could potentially imply both the study design and the data inconsistency: the reason may be that structural T1-weighted images alone are not enough to predict fluid intelligence scores; at the same time brain functional data like fMRI might have more predictive power for cognitive assessment.
The top performing model was a weighted average prediction of two VoxCNN neural networks trained on different parts of the training sample, highlighting the potential strength of the models’ ensembles yielded 70.635 MSE on the Validation set and 92.635 MSE on the Test set. Thus combination of different inputs, or so-called data fusion, gives us more information to built accurate prediction. Data fusion models are known to be successful in MRI segmentation applications, for example for epileptical foci detection [26].
6 Conclusion
In our work for the first time ensembles of VoxCNN networks were applied to the 3D brain imagery regression task. According to the results of this architecture we could consider it as a consistent predictive tool for large datasets with heavy and multi-modal inputs.
Due to the complex structure of the considered dataset there is enough room for further improvements. A future work on the model hyperparameters optimization is needed in order to achieve better network convergence. Advanced approaches to initialization of neural network parameters [16] and construction of ensembles [9] could be used. Sparse 3D convolutions could decrease memory requirements [36].
Transfer learning and domain adaptation techniques could potentially show better performance here [19, 25, 28]. Also it is possible to utilize multi-fidelity approaches when solving the regression problem with multi-modal data [13, 30, 31]. Conformal prediction framework [10, 17, 35] is a ready-to-use tool to assess prediction uncertainty.
References
Carroll, J.B.: Human Cognitive Abilities: A Survey of Factor-Analytic Studies. Cambridge University Press, Cambridge (1993)
RobertWCox: AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. In: Computers and Biomedical Research, vol. 29, no. 3, pp. 162–173 (1996)
Smith, S.M.: Fast robust automated brain extraction. Hum. Brain Mapp. 17(3), 143–155 (2002)
Avants, B.B., Tustison, N., Song, G.: Advanced normalization tools (ANTS). Insight j 2, 1–35 (2009)
Rohlfing, T., et al.: The SRI24 multichannel atlas of normal adult human brain structure. Hum. Brain Mapp. 31(5), 798–819 (2010)
Sadananthan, S.A., et al.: Skull stripping using graph cuts. NeuroImage 49(1), 225–239 (2010)
Avants, B.B., et al.: An open source multivariate framework for n-tissue segmentation with evaluation on public data. Neuroinformatics 9(4), 381–400 (2011)
Iglesias, J.E., et al.: Robust brain extraction across datasets and comparison with publicly available methods. IEEE Trans. Med. Imaging 30(9), 1617–1634 (2011)
Burnaev, E.V., Prikhod’ko, P.V.: On a method for constructing ensembles of regression models. Autom. Remote Control 74(10), 1630–1644 (2013)
Burnaev, E., Vovk, V.: Efficiency of conformalized ridge regression. In: Balcan, M.F., Feldman, V., Szepesvari, C. (eds.) Proceedings of the 27th Conference on Learning Theory. Proceedings of Machine Learning Research, PMLR, Barcelona, Spain, 13–15 Jun 2014, vol. 35, pp. 605–622 (2014)
Simonyan, K., Zisserman, A.: Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556 (2014)
Brown, S.A., et al.: The national consortium on alcohol and neurodevelopment in adolescence (NCANDA): a multisite study of adolescent development and substance use. J. Stud. Alcohol Drugs 76(6), 895–908 (2015)
Burnaev, E., Zaytsev, A.: Surrogate modeling of multifidelity data for large samples. J. Commun. Technol. Electron. 60(12), 1348–1355 (2015)
Szegedy, C., et al.: Going deeper with convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1–9 (2015)
Tompson, J., et al.: Efficient object localization using convolutional networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 648–656 (2015)
Burnaev, E., Erofeev, P.: The influence of parameter initialization on the training time and accuracy of a nonlinear regression model. J. Commun. Technol. Electron. 61(6), 646–660 (2016). ISSN 1555-6557
Burnaev, E., Nazarov, I.: Conformalized Kernel ridge regression. In: 2016 15th IEEE International Conference on Machine Learning and Applications (ICMLA), pp. 45–52 (2016)
Canziani, A., Paszke, A., Culurciello, E.: An analysis of deep neural network models for practical applications. arXiv preprint arXiv:1605.07678 (2016)
Goetz, M., et al.: DALSA: domain adaptation for supervised learning from sparsely annotated MR images. IEEE Trans. Med. Imaging 35(1), 184–196 (2016)
He, K., et al.: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on computer vision and pattern recognition, pp. 770–778 (2016)
Hosseini-Asl, E., Gimel’farb, G., El-Baz, A.: Alzheimer’s disease diagnostics by a deeply supervised adaptable 3D convolutional network. arXiv preprint arXiv:1607.00556 (2016)
Milletari, F., Navab, N., Ahmadi, S.-A.: V-net: fully convolutional neural networks for volumetric medical image segmentation. In: 2016 Fourth International Conference on 3D Vision (3DV), pp. 565–571. IEEE (2016)
Paul, E.J., et al.: Dissociable brain biomarkers of UID intelligence. NeuroImage 137, 201–211 (2016)
Szegedy, C., et al.: Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2818–2826 (2016)
Ghafoorian, M., et al.: Transfer learning for domain adaptation in MRI: application in brain lesion segmentation. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017. LNCS, vol. 10435, pp. 516–524. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66179-7_59
Hunyadi, B., et al.: Tensor decompositions and data fusion in epileptic electroencephalography and functional magnetic resonance imaging data. Wiley Interdiscip. Rev.: Data Min. Knowl. Discov. 7(1), e1197 (2017)
Korolev, S., et al.: Residual and plain convolutional neural networks for 3D brain MRI classification. In: IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), pp. 835–838. IEEE (2017)
Lu, H., et al.: When unsupervised domain adaptation meets tensor representations. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 599–608 (2017)
Pfefferbaum, A., et al.: Altered brain developmental trajectories in adolescents after initiating drinking. Am. J. Psychiatry 175(4), 370–380 (2017)
Zaytsev, A., Burnaev, E.: Large scale variable fidelity surrogate modeling. Ann. Math. Artif. Intell. 81(1), 167–186 (2017). ISSN 1573-7470
Zaytsev, A., Burnaev, E.: Minimax approach to variable fidelity data interpolation. In: Singh, A., Zhu, J. (eds.) Proceedings of the 20th International Conference on Artificial Intelligence and Statistics, Proceedings of Machine Learning Research, PMLR, Fort Lauderdale, FL, USA, 20–22 Apr 2017, vol. 54, pp. 652–661 (2017)
Chen, H., et al.: VoxResNet: deep voxelwise residual networks for brain segmentation from 3D MR images. NeuroImage 170, 446–455 (2018)
Hagler, D.J., et al.: Image processing and analysis methods for the adolescent brain cognitive development study. bioRxiv, p. 457739 (2018)
Ivanov, S., et al.: Learning connectivity patterns via graph kernels for fMRI-based Depression Diagnostics. In: Proceedings of IEEE International Conference on Data Mining Workshops (ICDMW), pp. 308–314 (2018)
Kuleshov, A., Bernstein, A., Burnaev, E.: Conformal prediction in manifold learning. In: Gammerman, A., et al. (eds.) Proceedings of the Seventh Workshop on Conformal and Probabilistic Prediction and Applications, Proceedings of Machine Learning Research, PMLR, vol. 91. pp. 234–253 (2018)
Notchenko, A., Kapushev, Y., Burnaev, E.: Large-scale shape retrieval with sparse 3D convolutional neural networks. In: van der Aalst, W.M.P., et al. (eds.) AIST 2017. LNCS, vol. 10716, pp. 245–254. Springer, Cham (2018). https://doi.org/10.1007/978-3-319-73013-4_23
Pominova, M., et al.: Voxelwise 3D convolutional and recurrent neural networks for epilepsy and depression diagnostics from structural and functional MRI Data. In: 2018 IEEE International Conference on Data Mining Workshops (ICDMW), pp. 299–307. IEEE (2018)
Sharaev, M., et al.: MRI-based diagnostics of depression concomitant with epilepsy: in search of the potential biomarkers. In: Proceedings of IEEE 5th International Conference on Data Science and Advanced Analytics, pp. 555–564 (2018)
Sharaev, M., et al.: Pattern recognition pipeline for neuroimaging data. In: Pancioni, L., Schwenker, F., Trentin, E. (eds.) ANNPR 2018. LNCS (LNAI), vol. 11081, pp. 306–319. Springer, Cham (2018). https://doi.org/10.1007/978-3-319-99978-4_24
Zhu, M., Liu, B., Li, J.: Prediction of general fluid intelligence using cortical measurements and underlying genetic mechanisms. In: IOP Conference Series: Materials Science and Engineering, vol. 381, no. 1, p. 012186. IOP Publishing (2018)
Eckle, K., Schmidt-Hieber, J.: A comparison of deep networks with ReLU activation function and linear spline-type methods. Neural Netw. 110, 232–242 (2019)
Acknowledgements
The work was supported by the Russian Science Foundation under Grant 19-41-04109.
The considered problem was formulated in the scope of the Project “Machine Learning and Pattern Recognition for the development of diagnostic and clinical prognostic prediction tools in psychiatry, borderline mental disorders, and neurology”, granted by Skoltech Biomedical Initiative Program, Skolkovo Institute of Science and Technology, Moscow, Russia.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Pominova, M. et al. (2019). Ensemble of 3D CNN Regressors with Data Fusion for Fluid Intelligence Prediction. In: Pohl, K., Thompson, W., Adeli, E., Linguraru, M. (eds) Adolescent Brain Cognitive Development Neurocognitive Prediction. ABCD-NP 2019. Lecture Notes in Computer Science(), vol 11791. Springer, Cham. https://doi.org/10.1007/978-3-030-31901-4_19
Download citation
DOI: https://doi.org/10.1007/978-3-030-31901-4_19
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-31900-7
Online ISBN: 978-3-030-31901-4
eBook Packages: Computer ScienceComputer Science (R0)