Abstract
Segmenting pancreas from abdominal CT scans is an important prerequisite for pancreatic cancer diagnosis and precise treatment planning. However, automated pancreas segmentation faces challenges posed by shape and size variances, low contrast with regard to adjacent tissues and in particular negligibly small proportion to the whole abdominal volume. Current coarse-to-fine frameworks, either using tri-planar schemes or stacking 2D pre-segmentation as prior to 3D networks, have limitation on effectively capturing 3D information. While iterative updates on region of interest (ROI) in refinement stage alleviate accumulated errors caused by coarse segmentation, extra computational burden is introduced. In this paper, we harness 2D networks and 3D features to improve segmentation accuracy and efficiency. Firstly, in the 3D coarse segmentation network, a new bias-dice loss function is defined to increase ROI recall rates to improve efficiency by avoiding iterative ROI refinements. Secondly, for a full utilization of 3D information, dimension adaptation module (DAM) is introduced to bridge 2D networks and 3D information. Finally, a fusion decision module and parallel training strategy are proposed to fuse multi-source feature cues extracted from three sub-networks to make final predictions. The proposed method is evaluated in NIH dataset and outperforms the state-of-the-art methods in comparison, with mean Dice-Sørensen coefficient (DSC) of 85.22%, and with averagely 0.4 min for each instance.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Pancreatic cancer, the main cause of several distinct neoplasms in the gland, is the seventh most common cause of cancer death [8]. Accurate pancreas segmentation is vital to effective detection of pancreatic cancer and more accurate treatment planning [3]. Nevertheless, since pancreas have complex anatomical structures, high diversity of shapes and negligibly small ratio with regard to abdominal volume, automated segmentation of pancreas from volumetric CT images remains a challenging task.
More recently, research efforts have been devoted to automated pancreas segmentation with convolutional neural networks [3, 6, 9,10,11,12]. The coarse-to-fine segmentation frameworks [10,11,12] assembling networks for coarse ROIs segmentation first and then further refinements, have been proposed to tackle the challenges introduced by the small ratio of pancreas in contrast to the whole abdominal CT volume. However, since the coarse segmentation network may not be able to produce a complete ROI initialization for the refining network, the coarse ROIs are often fed into refining network and updated according to current refining results in several iterators [10, 11]. While these iterative ROI refinements further improve the segmentation accuracy, the computational cost would be remarkably increased.
Due to its memory and computational efficiency, 2D pre-trained networks are often utilized to improve performance of pancreas segmentation [3, 6]. However, the lack of capturing the third dimensional information limits the segmentation performance of 2D networks. Consequently, tri-planar schemes [6, 10, 11] have been proposed to capture features along three orthogonal planes. However, features captured in such way cannot fully utilize 3D contents. Method in [9] tries to stack tri-planar predictions and forward them into a 3D network for further capturing of 3D features. However, the cascade of 2D and 3D networks is time-consuming and the independent training of 2D and 3D networks cannot be effectively harnessed for information propagation.
In this paper, we propose an improved coarse-to-fine framework to better utilize 3D information for segmenting small tissue/organ with high computational efficiency. To avoid additional computational cost introduced by iterative ROI refinements, we present bias-dice to 3D coarse segmentation network to raise the recall rate. In the refinement network, we utilize all pre-trained 3D encoder of coarse segmentation network and pre-trained 2D networks, specializing in capturing intra-slice features, to enhance predicted results. The dimension adaptation module (DAM) is proposed to capture interdependencies of adjacent slices. The fusion decision module and parallel training strategy are proposed to effectively fuse aforementioned multi-source features. We evaluate our method in the dataset of NIH [5] and obtain state-of-the-art results.
An illustration of our framework. (a) 3D coarse segmentation network trained with bias-dice loss proposes ROIs with high recall rates. (b) Refinement network that uses DAMs to transform intra-features of pre-trained networks into 3D and the fusion decision module to combine multi-source features to make accurate predictions.
2 Methods
As illustrated in Fig. 1, our framework is composed of 3D lightweight coarse segmentation network and multi-source refinement architecture. In coarse segmentation, bias-dice loss for the 3D U-Net is designed for optimal ROI extraction. In the refinement stage, the dimension adaptation module (DAM) is introduced to mine 3D information from pre-trained 2D networks. Multi-source 3D features are deconvoluted by 3D decoders for fusion decision module to make predictions.
2.1 3D Coarse Segmentation Network
A lightweight 3D U-Net [2], containing a 3D encoder to capture deep content features and a 3D decoder to make end-to-end classifications, is utilized for coarse segmentation. The output from decoder is fed into a decision module, composing of a 3D convolution layer and a sigmoid layer for generating probability map. From this probability map, ROI is defined as the bounding box containing the biggest connective region as the output of this coarse segmentation stage.
To reduce subsequent computational cost due to iterative updates on ROI definitions in the current coarse-to-fine methods [10, 11], we define a new bias-dice loss function to increase recall rates and thereby to alleviate incomplete ROI definitions. The bias-dice loss function is defined as below:
Where \(p_{i}\) denotes predicted probability map and \(g_{i}\) is the ground truth, \(\epsilon \) is smoothness term to avoid the risk of division by 0. Actually, the item of \(\sum _{i=1}^{N}{p_{i}(1-g_{i})}\), \(\sum _{i=1}^{N}{g_{i}p_{i}}\) and \(\sum _{i=1}^{N}{g_{i}(1-p_{i})}\) are respectively soft items of TP, FP and FN. Compared to original dice loss, which sets \(\beta =1\) in the soft FN item of \(\sum _{i=1}^{N}{g_{i}(1-p_{i})}\), the bias-dice loss emphasizes importance of recall and sets \(\beta >1\) (\(\beta =3\) in this paper) to introduces penalty on FN and ensure high recall rates.
2.2 Dimension Adaptation Module for Aggregating 3D Features
2D networks can be effectively initialized by pre-trained models to capture powerful intra-slice features from volumetric CT images. However, only using 2D networks to segment pancreas neglects important information along z-axis, which therefore limits the segmentation performance. In order to utilize pre-trained 2D networks to extract intra-slice features without missing of inter-slice features, we firstly transform 2D pre-trained networks into 3D for direct prediction of volumetric images and propose DAMs to aggregate contexts along z-axis.
Transforming 2D Layers into 3D Layers: As shown in the Fig. 2(a), a 2D convolution layer with kernel of (h, w) can be equivalently expressed as a 3D convolution layer with kernel of (1, h, w). Similarly, the 2D pooling operations can be transformed into the corresponding 3D pooling layers. So we can replace 2D convolution and 2D max-pooling layers with 3D layers to transform 2D pre-trained networks, such as Vgg16 and Res18, into 3D versions to directly obtain intra-slice features of a volumetric image.
Dimension Adaptation Module to Capture 3D Features: Aforementioned intra-slice features can be fed into DAMs to capture inter-slice features. The architecture of DAM is shown in Fig. 2(b), in which, the first 3D convolution layer compresses channel number of intra-slice features by eight times to relieve memory cost. And the following convolution layers with kernel of (3, 3, 3) and max-pooling layers, which only compress features along z-axis to broaden receptive field, are utilized to mine relationship of adjacent 2D sectional slices.
2.3 Parallel Training Strategy and Inferencing
Parallel Training Strategy. In the refinement stage, to effectively capture 3D features, 3D encoder of coarse segmentation network, Vgg16 accompanied with DAMs and Res18 accompanied with DAMs, are simultaneously adopted. To effectively utilize them and unify multi-source cues, we propose a parallel training strategy. As shown in Fig. 3, we respectively set three decoders and add three extra decision modules supplement supervisions of the conventional fusion decision module for the multi-source features. The updated loss is defined as below:
Where \(Loss_{i}\) denotes the additional supervision of the i-th decision module and \(Loss_{fused}\) is the loss of fusion decision module. N is the number of sub-networks and is 3 in refining network. All of these losses are dice loss.
From Whole Volumes to ROIs. To effectively utilize all data, we first respectively train three sub-networks on the whole data, and then we integrate pre-trained models into a unified one. Then, we further train the total network on the data of ROIs by parallel training strategy.
For the training of networks on whole volumes, we forward \(64\times 192 \times 192\) volume images into networks at batch size of 2 each time. For the training of networks based on pre-trained 2D networks, we first freeze encoders and warm up decoders and DAMs with Adam with \(lr=0.001\). Then, we fine-tune encoders with \(lr=0.0001\). We divide 8 instances as validation data, so early stopping can be conducted to stop training if validation loss does not improve in 10 epochs. The max epoch is 200 and networks are updated 60 times each epoch.
For the training on the data of ROIs, we put a whole volumetric ROI into networks every time. We first spend 5 epochs to warm up fusion decision module. Then the lr is set as 0.0001 for Vgg16 and Res18, while other parts with the \(lr=0.001\). If the validation loss does not decrease in 10 epochs, we will set \(lr=0.0001\) for all networks, meanwhile, early stopping strategy is conducted to stop training after 10 epochs without improvement.
Inferencing. The region of abdomen is firstly segmented from the whole volumetric CT image using OTSU [4] to shrink the searching region. Then, the volumetric image of abdomen is fed into the coarse segmentation network to obtain initial results, which will be further binarized by a threshold value of 0.1. The biggest connective region of binary result will be set as ROI, and the margins of ROI region will be extended by 8, 32, 32 on z, y, x axis to provide sufficient spatial contexts. Finally, the extended ROI will be entered into the refinement segmentation network to get fine results. The predicted results will be binarized by 0.5 and the biggest connective region will be the final segmentation result.
3 Experiments and Discussion
3.1 Dataset and Evaluation Metrics
Experiments are conducted on the NIH pancreas segmentation dataset [5], which contains 82 abdominal CT volumes. The size of each CT scan is \(512\times {512}\times {D}\), where D ranges from 181 to 466. We perform 4-fold cross-validation. Every CT is evaluated by computing the DSC: \(DSC=\frac{2{|P\times {G}|}}{|P|+|G|}\), where P is the binary results of predictions and G is the ground truth.
3.2 Results
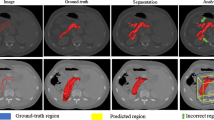
We compare our proposed method with extensive state-of-the-art methods. The DSC values and inference times of different methods are shown in Table 1. In addition to presenting the results of single fusion decision module, we also show the results of integrating all four decision modules using average operation.
According to results shown in Table 1, our proposed method has the better mean DSC, where the mean DSC is \(85.09\%\) for fusion decision module and is \(85.22\%\) for integrated one. And the worst instance can still have a DSC of \(71.42\%\), which shows the good robustness of the proposed method. What’s more, the method proposed is much more time-effective with only about 0.4 min to realize the segmentation of one single instance.
3.3 Discussion
The Contribution of Bias-Dice: As shown in Fig. 4, coarse segmentation method based on bias-dice loss has a better performance in the average ROI recall with \(99.1\%\) and mean DSC with \(79.4\%\) than based on original dice loss. The bias-dice makes network pay more attention on recall rate, therefore, the results have less FNs, which reduces the missing of meaningful pancreas regions.
The Contribution of Parallel Training: Both results of methods with and without parallel training are shown in Table 1. Compared to method without parallel training with mean DSC of \(83.99\%\) and minimum DSC of \(66.62\%\), method with parallel training can be much more stable and effective with mean DSC of \(85.09\%\) and minimum DSC of \(71.42\%\). The utilization of parallel training can effectively alleviate over-fitting caused by massive parameters and improve the performance of unified network.
The Optimal “Speed-Accuracy Tradeoff”: Our method provides an optimal “speed-accuracy tradeoff” with both improved accuracy and reduced inference time in comparison to the state-of-the-art methods with best accuracy and fastest speed. Compared with the method with highest mean DSC, our method remarkably accelerates inference from 1.4 min to 0.44 min. In comparison with the fastest method, our method increases mean DSC from \(82.5\%\) to \(85.2\%\) and further reduces inference time from 0.9 min to 0.44 min.
4 Conclusion
In this paper, we improve the coarse-to-fine framework for faster and more accurate pancreas segmentation. In the coarse segmentation stage, our proposed bias-dice loss function adds a penalty in the soft FN item to achieve high recall rates of ROIs, and to effectively alleviate missing informative regions without needs for iterative ROI adjustments. In the refinement stage, the DAMs, bridging pre-trained 2D networks and 3D features, is presented to capture inter-slice features from pre-trained 2D networks. Further, a fusion decision module and parallel training strategy are introduced to effectively train the three sub-networks in a unified manner. The experimental results demonstrate that our proposed method outperforms the current methods based on convolutional neural networks in comparison and achieved highest computational efficiency with about 0.4 min per instance.
References
Cai, J., Lu, L., Zhang, Z., Xing, F., Yang, L., Yin, Q.: Pancreas segmentation in MRI using graph-based decision fusion on convolutional neural networks. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 442–450. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_51
Falk, T., et al.: U-Net: deep learning for cell counting, detection, and morphometry. Nat. Methods 16(1), 67 (2019)
Farag, A., Lu, L., Turkbey, E., Liu, J., Summers, R.M.: A bottom-up approach for automatic pancreas segmentation in abdominal CT scans. In: Yoshida, H., Näppi, J., Saini, S. (eds.) ABD-MICCAI 2014. LNCS, vol. 8676, pp. 103–113. Springer, Cham (2014). https://doi.org/10.1007/978-3-319-13692-9_10
Otsu, N.: A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 9(1), 62–66 (1979)
Roth, H.R., et al.: DeepOrgan: multi-level deep convolutional networks for automated pancreas segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9349, pp. 556–564. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24553-9_68
Roth, H.R., Lu, L., Farag, A., Sohn, A., Summers, R.M.: Spatial aggregation of holistically-nested networks for automated pancreas segmentation. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 451–459. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_52
Roth, H.R., et al.: Spatial aggregation of holistically-nested convolutional neural networks for automated pancreas localization and segmentation. Med. Image Anal. 45, 94–107 (2018)
Stewart, B., Wild, C.P., et al.: World cancer report 2014 (2014)
Xia, Y., Xie, L., Liu, F., Zhu, Z., Fishman, E.K., Yuille, A.L.: Bridging the gap between 2D and 3D organ segmentation with volumetric fusion net. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11073, pp. 445–453. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00937-3_51
Yu, Q., Xie, L., Wang, Y., Zhou, Y., Fishman, E.K., Yuille, A.L.: Recurrent saliency transformation network: incorporating multi-stage visual cues for small organ segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 8280–8289 (2018)
Zhou, Y., Xie, L., Shen, W., Wang, Y., Fishman, E.K., Yuille, A.L.: A fixed-point model for pancreas segmentation in abdominal CT scans. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017. LNCS, vol. 10433, pp. 693–701. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66182-7_79
Zhu, Z., Xia, Y., Shen, W., Fishman, E., Yuille, A.: A 3D coarse-to-fine framework for volumetric medical image segmentation. In: 2018 International Conference on 3D Vision (3DV), pp. 682–690. IEEE (2018)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Chen, H., Wang, X., Huang, Y., Wu, X., Yu, Y., Wang, L. (2019). Harnessing 2D Networks and 3D Features for Automated Pancreas Segmentation from Volumetric CT Images. In: Shen, D., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2019. MICCAI 2019. Lecture Notes in Computer Science(), vol 11769. Springer, Cham. https://doi.org/10.1007/978-3-030-32226-7_38
Download citation
DOI: https://doi.org/10.1007/978-3-030-32226-7_38
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-32225-0
Online ISBN: 978-3-030-32226-7
eBook Packages: Computer ScienceComputer Science (R0)