Abstract
In this paper, we propose a new weakly supervised 3D brain lesion segmentation approach using attentional representation learning. Our approach only requires image-level labels, and is able to produce accurate segmentation of the 3D lesion volumes. To achieve that, we design a novel dimensional independent attention mechanism on top of the Class Activation Maps (CAMs), which refines the 3D CAMs to obtain better estimates of the lesion volumes, without introducing significantly more trainable variables. The generated attentional CAMs are then used as a source of weak supervision signals to learn a representation model, which can reliably separate the voxels belong to the lesion volumes from those of the normal tissues. The proposed approach has been evaluated on the publicly available BraTS and ISLES datasets. We show with comprehensive experiments that our approach significantly outperforms the competing weakly-supervised methods in both initial lesion localization and the final segmentation, and is able to achieve comparable Dice scores in segmentation comparing to the fully supervised baselines.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Magnetic Resonance Imaging (MRI) has become one of the key diagnostic techniques for brain lesion analysis, offering a non-intrusive and radiation-free modality to detect abnormalities. For many diseases such as brain tumor, the precise identification and segmentation of the lesion volumes is a fundamental prerequisite for further treatments including surgery or radiation therapy. Traditionally, this process is performed by the experienced medical experts, which is expensive, tedious and more importantly prune to the inter-rater/test-retest variability [11]. Recently, there has been a solid body of work on automatic lesion segmentation [5, 8, 13], offering a faster and more objective alternative to human labor. In particular, the deep learning based methods have demonstrated impressive performance in various segmentation tasks, and are robust enough to process lesion volumes with irregular shapes or challenging sizes [8, 13]. However, most of the existing deep learning methods are supervised, whose success depends on a substantial amount of training data with accurate labels. Particularly for the task of brain lesion segmentation, such labels have to be the accurate lesion boundaries in the 3D space, which are often annotated by the experts manually. They are extremely expensive and time-consuming to obtain, which significantly limits the application of those approaches in clinical practice.
On the other hand, weakly supervised segmentation approaches which only require the coarse-grained (e.g. image-level) labels point to a promising direction [3, 4, 6, 7, 10]. These approaches aim to exploit the minimum level of expert annotations, while being able to generate the fine-grained segmentation automatically. Many of these weakly supervised approaches leverage the Class Activation Maps (CAMs) [14] as a key step for segmentation. For example, the work in [7] uses multi-layer CAMs to detect histological features of glioma in CLE images, while [3] proposes an iterative mining pipeline to localise lesion in different areas. However, a major drawback of CAMs is that they can be very noisy in practice, and thus it is often challenging to derive good initial segmentation proposals from them, especially for 3D images.
In this paper, we propose a novel weakly supervised segmentation approach for brain lesion in MRI images, which only requires image-level binary labels indicating whether the lesion is present or not. Such weak labels are much easier to obtain in practice, and thus our approach can generate high quality segmentation proposals on the vast unlabeled data with minimum human input, which can then be refined by experts to produce accurate voxel-level labels in 3D. In particular, we develop a new dimensional independent attention mechanism on top of the standard CAMs, which can significantly improve lesion localization without introducing excessive trainable variables to the network. This attentional CAM provides high quality estimates of the lesion regions as well as normal tissues, which are then used to train a representation model. The learned model can reliably distinguish the voxels of lesion volumes from those belonging to the normal tissues, and thus generate the accurate lesion segmentation. We evaluate the proposed approach on the public BraTS [2] and ISLES [9] datasets. Extensive experiments show that with the proposed attentional CAMs, our approach is able to acquire much better initial estimates of lesion regions compared to the competing methods, and the proposed representation learning technique is evidently beneficial in improving segmentation performance, significantly outperforming the state-of-the-art while offering comparable accuracy with the fully supervised baselines.
2 Method
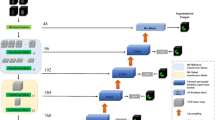
The proposed weakly supervised 3D lesion segmentation approach consists of three steps: (i) obtaining the initial lesion regions with image-level labels; (ii) learning a representation model to distinguish the voxels belong to normal and lesion tissues; and (iii) using the learned model to obtain the fine-grained voxel-level segmentation of lesion volume. Figure 1 shows the overview of the proposed approach. In particular, our approach is based on two deep neural networks: (a) a classification network which is used to compute the 3D Class Activation Maps (CAMs); and (b) another network which learns the voxel representation model and performs the segmentation. The remainder of this section describes the design of the two networks and training techniques for them in more details.
2.1 Lesion Localization with Attentional CAM
Generating CAMs with Image-Level Labels: In many state of the art weakly supervised approaches, the Class Activation Maps (CAMs) [14] are often considered as the seeds for later segmentation. In the proposed approach, we also consider CAMs as the initial segmentation proposals, which can roughly localize the lesion volumes within the images. In particular, given the image-level labels i.e. binary labels indicating if lesion tissues exist, the standard technique [15] to obtain the CAM is to train a classification network with global average pooling (GAP) or global mean pooling (GMP) followed by a fully connected layer. The CAM of a positive sample (image with lesion) is computed by:
where \(\mathbf {w}\) is the weights associated to the positive class, F is the feature map extracted by the last convolutional layer before the GAP/GMP operation, and F(h, w, d) is the feature vector at location (h,w,d). In practice, C is typically normalized to make the maximum activation equals to one, and a threshold is often used to extract the region of target objects, which can be used as an initial proposal for segmentation.
Refining CAMs with Attention: However for 3D medical images, in some cases the lesion tissues may only occupy a small fraction of the entire volume, e.g., in our experiments with the ISLES [9] and BraTS datasets [2], the tumor or stroke volumes can be just 0.1% of the whole brain. For those samples, the CAMs tend to be very noisy, making it challenging to select the appropriate threshold to obtain the desired lesion regions. In practice, if GAP is used in the network then we typically need to consider larger activation values which make training more difficult to converge. On the other hand, if GMP is considered, the network may focus too much on small areas with stronger responses, and generate incomplete regions that won’t be able to cover the whole lesion volumes.
To address this problem, we proposed to use the attention mechanism to refine the generated CAMs. Conceptually this can be achieved by directly applying the learned attention sores to the 3D CAMs. However, in practice, it is prohibitively expensive for 3D images since the amount of trainable parameters are significantly larger than 2D, which can easily lead to over-fitting. Therefore, in this paper, we consider an approximation approach, which enforces attention along individual dimensions and fuses the re-weighted features afterwards. Concretely, let \(F\in \mathbb {R}^{H\times W \times D \times K}\) be the activation map extracted by the last convolutional layer. F can be viewed as K 3D feature cubes \(F_k \in \mathbb {R}^{H \times W \times D}\). For each cube \(F_k\), we first apply average pooling to its slices along the three different dimensions. For example along the dimension of size D, we do average for the D feature slices, each of which is a \(H\times W\) feature, and obtain a vector \(F^{D}\in \mathbb {R}^{D\times K}\) across the K cubes. We then consider a self-attention over this feature \(F^{D}\), where the attention scores \(s_d\) (\(1\le d \le D\)) is calculated as:
Here \(F^{D}(d)\) is the d-th feature in \(F^{D}\), and \(\mathbf {w}_{\text {att}}\) are the weights learned via training. The attention scores \(s_d\) are then used to re-weight the generated CAM along the dimension of size D, i.e. multiplied to the 2D slices of the activation values (with size \(H\times W\)) in the CAM: \(C'_{D}(d) = s_d \cdot C(d)\).
Similarly, we can learn the attention scores along the other two dimensions (with size H and W), which are used to obtain the re-weighted CAMs \(C'_{H}\) and \(C'_{W}\). Then the final attentional CAM (ACAM) \(C'\) is computed by fusing the three \(C_D\), \(C_H\) and \(C_W\), which is also normalized to [0, 1].
Intuitively, the learned attention scores specify how important the feature slices are with respect to the final classification results (lesion or not) along a certain dimension, which is exploited to refine the standard CAM. It is also worth pointing out that here we implicitly assume that the attention scores along different dimensions are independent, and thus they are actually approximations to the full 3D self-attention, which however would require much more training effort. In our experiment, we show that the proposed attentional CAM (ACAM) can achieve much better performance than the standard CAMs (using GMP or GAP), while won’t incur significantly expensive training costs.
2.2 Weakly Supervised Representation Learning for Segmentation
As discussed above, the ACAMs generated by the classification network provide rough estimates of the 3D lesion volumes. To obtain the accurate voxel-level segmentation, in this paper we use another network, which exploits the ACAMs as a source of weak supervision. The key idea is that although ACAMs are not globally accurate segmentation, in regions with high confidence they can still provide valuable information on features of the foreground (lesion) and background (normal tissues). We could then leverage that to learn a universal representation model, with which voxels from the foreground can be effectively separated from those from the background, and thus obtain the voxel-level segmentation.
Learning the Representation Model: To learn the representation model, we first randomly select the voxels with high confidence from both the foreground (lesion) and background (normal tissues) according to the ACAMs. In particular, we select a foreground voxel if the ACAM \(C'(h,w,d)\ge \theta _{FG}\), while the background if \(C'(h,w,d)\le \theta _{BG}\). In our experiments, we set \(\theta _{FG}\) to 0.1 and \(\theta _{BG}\) to 0.01 respectively.
With the selected set of high confidence voxels V, we aim to learn a representation model \(f_{R}\) in the form of a neural network, which maps an arbitrary voxels p to its embedding vector \(f_R(p)\). In this case, the distance between two voxels p and q can be defined as \(D(p,q) = (1 + \exp \{- \Vert f_R(p) - f_R(q)\Vert _2^2\})^{-1}\), where \(\Vert \cdot \Vert _2\) is the \(L_2\) norm. We train \(f_{R}\) using a cross-entropy loss:
where the indicator function \(\mathbbm {1}(p,q)\) returns 1 if the voxels p and q both belong to the foreground or background, while 0 if p and q are different.
Segmentation Based on Voxel Affinity: Let p be a foreground voxel in V. With the learned representation model \(f_R\), we could obtain a lesion segmentation candidate \(S_p\) by selecting all voxels q that are close enough (with high affinity) to p, i.e. \(S_p = \{q| D(p,q) \ge \lambda \}\), where the rest is considered as the background (normal tissues). In practice, the quality of the segmentation \(S_p\) depends on the selection of the seed voxel p, which can lead to noisy results in some cases. To mitigate that, in this paper, we firstly use multiple foreground seed voxels in V to generate an array of segmentation candidates \(\varvec{S}\). Then for each candidate \(S_p \in \varvec{S}\), we assign a confidence score \(c_p\), which is computed as the mean of the voxel ACAM values, i.e. the average confidence that the voxels in \(S_p\) belong to the foreground. Finally, we combine the segmentation candidates \(\varvec{S}\) by non-maximum suppression according to the confidence scores, and obtain the final segmentation of the lesion volume.
3 Evaluation
3.1 Experimental Setup
Datasets: We consider two datasets in our experiments: (i) the Multimodal Brain Tumor Segmentation dataset (BraTS 2017) [2] and (ii) the Ischemic Stroke Lesion Segmentation (ISLES 2017) dataset [9]. The BraTS dataset contains multimodal MRI images including T1, T1c, T2, and Flair, where the training set has 210 samples and the test set contains 47 samples. The ground truth segmentation includes four tumor tissue classes, but in this paper, we only consider those for the whole tumors. The ISLES dataset also contains multi-spectral MRI data of 43 stroke patients, with ground truth segmentation provided.
Competing Approaches: For initial lesion localization, we compare the proposed attentional CAM with CAM(GMP) and CAM(GAP), which are the standard approaches of generating CAMs using Global Maximum Pooling(GMP) and Global Average Pooling(GAP) respectively. For lesion segmentation we compare the proposed weakly supervised representation learning approach (ACAM+WSR) with: (i) a fully supervised UNet [12] (S-UNet) trained with the ground truth; and (ii) a weakly supervised UNet (WS-UNet), which uses the results of our ACAM as the weak supervision signals to train the UNet.
Metrics: We consider the Dice, Sensitivity and Specificity as the evaluation metrics, which are computed as: Dice = 2TP/(2TP+FP+FN); Sensitivity = TP/(TP+FN); and Specificity = TN/(TN+FP) respectively. Here TP, FP, TN, and FN are true positive, false positive, true negative and false negative.
Implementation: Our networks are implemented with Tensorflow [1], and trained on a single Titan X GPU with the Adam optimiser of learning rate 0.001. The network to generate ACAM is a VGG-like 3D convolutional network, where at the end of the conv layer we incorporate the proposed attention mechanism. The network for representation learning shares the similar structure with 3D UNet. We train the networks with only binary labels, i.e. if this image contains lesion or not, and report the segmentation performance with respect to the ground truth segmentation labels. For BraTS dataset, we report results returned from the official evaluation server, while for the ISLES dataset, we randomly divide training and testing sets (3:1) and report results on the testing set.
3.2 Results
Lesion Localization: Table 1 shows the lesion localisation performance of the competing approaches. We can see that on the BraTS dataset, the standard CAM with GMP has the much lower Dice and Specificity scores (around 0.32 and 0.27), which won’t be able to localise the lesion volume. The CAM(GAP) works better than CAM(GMP), but it is the proposed ACAM that achieves the best results overall (up to 42% improvement in Dice and 46% in Sensitivity). It confirms that the attention mechanism can help the network to focus more on the lesion volumes, and significantly improve the localization performance (see Fig. 2(b) and (c)). On the other hand, we see that on ISLES dataset the gap between CAM(GMP) and CAM(GAP) is smaller, where CAM(GMP) even has a slightly better Dice score. This is because unlike in the BraTS dataset, the lesion volumes in ISLES dataset tend to be smaller, where the max pooling used in CAM(GMP) is more suitable to detect such small regions. Again we see the proposed ACAM achieves the best results, offering up to 6% improvement in Dice and 10% better Sensitivity scores.
Lesion Segmentation: We are now in a position to evaluate the performance of lesion segmentation of the competing approaches. Table 2 shows the results on both BraTS and ISLES datasets. Note that here the S-UNet is a fully supervised approach, which has access to the ground truth segmentation. On the other hand, both WS-UNet and the proposed ACAM+WSR only use the image level labels. We see that the proposed ACAM+WSR generally outperforms the weakly supervised WS-UNet, e.g., in the ISLES dataset it enjoys roughly 5% better dice and 10% better sensitivity scores, while in the BraTS the improvements are 3% and 12.5% respectively. This means the representation learning does help to effectively extract lesion volumes from the normal tissues, leading to more accurate segmentation. On the other hand, we see that the gap between the proposed ACAM+WSR and the fully supervised S-UNet is only about 6% in Dice (note that for ACAM+UNet the gap is 11%), which means even with image-level labels, our weakly supervised approach could achieve comparable accuracy with fully supervised baseline (see Fig. 2(d) and (e) for an example).
4 Conclusion
In this paper, we present a new weakly supervised brain lesion segmentation approach that only requires image-level labels to generate the accurate voxel-level 3D boundaries of the lesion volumes. We develop a novel attentional CAM technique to better localize the lesion regions, and exploit representation learning to further improve lesion segmentation. The proposed approach has been validated on two public datasets, the BraTS and ISLES, where experimental results show that our approach offers superior performance to the state-of-the-art, and can achieve comparable segmentation accuracy with the fully supervised methods. For future work, we would like to explore approaches that can detect multiple classes of the lesion structure with similar weak supervision signals and consider more types of lesion data.
References
Tensorflow. https://www.tensorflow.org. Accessed 01 Apr 2019
Bakas, S., et al.: Advancing the cancer genome atlas glioma mri collections with expert segmentation labels and radiomic features. Sci. Data 4, 170117 (2017)
Cai, J., Lu, L., Harrison, A.P., Shi, X., Chen, P., Yang, L.: Iterative attention mining for weakly supervised thoracic disease pattern localization in chest x-rays. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11071, pp. 589–598. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00934-2_66
Cai, J., et al.: Accurate weakly-supervised deep lesion segmentation using large-scale clinical annotations: slice-propagated 3D mask generation from 2D RECIST. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11073, pp. 396–404. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00937-3_46
Despotović, I., Goossens, B., Philips, W.: MRI segmentation of the human brain: challenges, methods, and applications. Computat. Math. Methods Med. 2015 (2015)
Hwang, S., Kim, H.-E.: Self-transfer learning for weakly supervised lesion localization. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 239–246. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_28
Izadyyazdanabadi, M., et al.: Weakly-supervised learning-based feature localization for confocal laser endomicroscopy glioma images. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11071, pp. 300–308. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00934-2_34
Kamnitsas, K., et al.: Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 36, 61–78 (2017)
Maier, O., et al.: ISLES 2015-a public evaluation benchmark for ischemic stroke lesion segmentation from multispectral MRI. Med. Image Anal. 35, 250–269 (2017)
Mlynarski, P., Delingette, H., Criminisi, A., Ayache, N.: Deep learning with mixed supervision for brain tumor segmentation. arXiv preprint arXiv:1812.04571 (2018)
Porz, N., et al.: Multi-modal glioblastoma segmentation: man versus machine. PLoS ONE 9(5), e96873 (2014)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Tseng, K.L., Lin, Y.L., Hsu, W., Huang, C.Y.: Joint sequence learning and cross-modality convolution for 3D biomedical segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 6393–6400 (2017)
Zhou, B., Khosla, A., Lapedriza, A., Oliva, A., Torralba, A.: Learning deep features for discriminative localization. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2921–2929 (2016)
Zhou, B., Khosla, A., Lapedriza, A., Oliva, A., Torralba, A.: Learning deep features for discriminative localization. In: CVPR, pp. 2921–2929 (2016)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Wu, K., Du, B., Luo, M., Wen, H., Shen, Y., Feng, J. (2019). Weakly Supervised Brain Lesion Segmentation via Attentional Representation Learning. In: Shen, D., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2019. MICCAI 2019. Lecture Notes in Computer Science(), vol 11766. Springer, Cham. https://doi.org/10.1007/978-3-030-32248-9_24
Download citation
DOI: https://doi.org/10.1007/978-3-030-32248-9_24
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-32247-2
Online ISBN: 978-3-030-32248-9
eBook Packages: Computer ScienceComputer Science (R0)