Abstract
Human brain networks convey important insights in understanding the mechanism of many mental disorders. However, it is difficult to determine a universal optimal among various tractography methods for general diagnosis tasks. To address this issue, tentative studies, aiming at the identification of some mental disorders, make an effective concession by exploiting multi-modal brain networks. In this paper, we propose to predict the clinical measures as a more comprehensive and stable assessment of brain abnormalities. We develop a graph convolutional network (GCN) framework to integrate heterogeneous brain networks. Particularly, an adaptive pooling scheme is designed, catering to the modal structural diversity and sharing the advantages of locality, loyalty and likely as in standard convolutional networks. The experimental results demonstrate that our method achieves state-of-the-art prediction results, and validates the advantages of the utilization of multi-modal brain networks in that, more modals are always at least as good as the best modal, if not better.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
1 Introduction
Large-scale connection in the brains convey important insights in understanding the underlying yet unknown mechanism of many mental disorders [3, 7, 16]. With whole brain tractography, brain’s anatomical networks represented as major fiber bundles can be reconstructed from diffusion-weighted MRI (DWI). There are various brain networks, generated from different tractography algorithms based on either voxel-wise diffusion model or cross-voxel fiber tracking, each finding the place in revealing targeted brain abnormalities, such as autism spectrum disorder [12], Parkinson’s disease [4], and even in genetics. Nevertheless, for distinctive diagnosis tasks it is elusive to decide a universally optimal method and accompanied processing, e.g. dimension reduction [19], as that these tractography algorithms differ in the selection and accuracy of fiber extraction, robustness, and particularly the relevance between the extracted fiber bundles and the tasks. Essentially, tentative studies have demonstrated that multi-modal brain networks can provide complementary viewpoint toward the classification tasks, in leveraging the scattered information from acquisitions with diverse tractography algorithms. For example, it is showed that multi-view graph convolutional network [20] has state-of-the-art performance in classifying Parkinson’s disease (PD) status.
To take one step further, we propose to predict the clinical measures, instead of directly classifying the disease status. The behind motivation lies in that, many mental disorders are degenerative, which can be inferred from the gradual progress of brain connectivity patterns, and that clinical measures, compared to simply classification, better capture the progress. Through integrating multi-modal brain networks in our prediction, a comprehensive assessment is constructed, and the potential deterioration from sub-optimal of single tractography is alleviated. To address the prediction problem, we resort to a cascade model, composed of a heterogeneous graph convolutional network (GCN) for brain network embeddings and a multi-layer perceptron (MLP) for regression. Our contributions are two-folded: first, we propose a heterogeneous GCN to predict the clinical scores from multi-modal brain networks, which benefits from the natural graph structures of diverse brain networks; second, an adaptive pooling scheme, driven by both graph structure and network patterns, is proposed, which is beneficial from gathering local information, yielding a faithful graph with smaller size, and enjoying efficiency in both computation and training. We name the proposed method as “heterogeneous” in that the graph convolution and pooling are customized for varied modal. The proposed method is verified on the data from the Parkinson Progression Marker Initiative (PPMI) [13], a cohort study aiming at identifying and validating PD progression markers. The experimental results show that our method outperforms related baselines significantly. It is also demonstrated that by integrating multi-modal brain networks, the proposed method achieves higher accuracy, and yields more stable prediction.
The rest of this paper is organized as follow. Sect. 2 provides the preliminary and describes the detail of the proposed method. Sect. 3 shows the experiments and the results. Sect. 4 concludes the paper.
2 Methodology
2.1 Preliminary
A graph can be represented as \(\{ V,E,W\}\), with \(V = \{ v_1, v_2,\cdots , v_n\}\) the set of n vertices, \(E \subseteq V \times V\) the set of m edges, and \(W \in \mathcal {R}^{n\times n}\) the weighted adjacency matrix of the graph. In this paper, vertices are Region Of Interest (ROIs). Graph Laplacian is an operation in spectral graph analysis, typically defined as \(L_c = D-W\) in combinatorial form and \(L_n = I_n- D^{-1/2}WD^{-1/2}\) in normalized form, with \(D \in \mathcal {R}^{n\times n}\) the diagonal matrix and \(I_n\) an identity matrix. Graph convolutional network (GCN) [5] is designed as an extension of convolutional neural network (CNN), to analysis the signals on nodes, with a given graph structure. One strategy is to conduct graph convolution in frequency domain using the eigenvalue decomposition of graph Laplacian, \(L = U^T \varLambda U\), and \(U = [u_0,u_1,\cdots ,u_{n-1}]\) specifies a Fourier basis. The graph Fourier transform [15] is then defined as \(\hat{x} = U^Tx\), with \(x\in \mathcal {R}^{n}\) the signal, and the inverse transform \(x = U\hat{x}\). The spectral representation of node signals, \(\hat{x}\), allows the fundamental filtering operation for graphs. For computational accessibility, polynomial parametrization for localized filters [5] is proposed through learning the coefficients \(\varTheta _K\) of a K-order Chebyshev polynomial. The filter is defined as \(g_{\theta }(\varLambda ) = \sum _{k=0}^{K-1}{\theta _k T_k(\varLambda )}\), and the graph convolution is defined as \(y = Ug_{\theta }(\varLambda )U^Tx\), with y the filtered signal, \(\theta _k\) the trainable parameters and \(T_k(\varLambda )\) the polynomials. Parallel to CNN, pooling operation in graph settings is accomplished by the graph coarsening [6] procedure. By truncating the Chebyshev polynomial to only first order, a faster version of GCN can be reformulated [11] similar to multi-layer perceptron, with each layer defined as \(y = \sigma (\tilde{D}^{-1/2}\tilde{A}\tilde{D}^{-1/2}x\varTheta )\), with \(\tilde{A} = A+I_n\), \(\tilde{D}\) accordingly defined as D, \(\varTheta \) the trainable parameters, and \(\sigma (\cdot )\) an activation function.
In graph classification, a major concern is to represent graphs with embeddings. Previous approaches [18] include averaging all the node embeddings in a final layer, computing “virtual node” connected to all nodes, operating over sets using deep node aggregation, concatenating all embeddings, and training hierarchical structure. A majority of these methods apply a deterministic graph clustering subroutine, while some end-to-end [18, 21] methods require additional structure to compute the pooling structure.
At last we want to include a short discussion on the relation of graph convolution and traditional methods. Essentially GCN is a flexible and mixed model of classical methods: it is rooted in graph spectral theory, in the manner the node signals are analyzed; meanwhile, the convolution is closely related and can be re-formulated as random-walk [8] or Weisfeiler-Lehman Method [17].
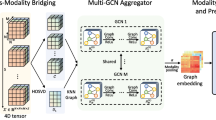
The proposed heterogeneous GCN for PD clinical scores prediction. (a) illustrates the entire structure, in which multi-modal brain networks are generated from MRIs, and sequentially processed by GCN and MLP; (b) depicts the stacked convolutional layer and pooling layer; (c) provides a detailed description of the pooling procedure, including node merge, graph distillation and feature pooling.
2.2 Predicting PD Clinical Scores via Heterogeneous GCN
The proposed method has two stages, as illustrated in Fig. 1. In the first stage, the per-modal embeddings for brain networks are generated via heterogeneous GCN; in the second, the concatenated embeddings are regressed to the clinical scores via MLP. Parallel to convolutional neural networks, the proposed GCN is formed by stacking graph convolutional layer and pooling layer sequentially. The fast graph convolutional [11] is applied, using corresponding rows of brain network matrix as node features. We also propose a novel efficient adaptive pooling scheme to learn data-driven pooling windows, which in turns can construct reduced while structural-preserving graphs and aggregate pooled features.
Although graph convolution is a quite established technique, pooling on graphs is challenging in many senses. A major difficulty is the structural irregularity of graph data, compared to naive application scenarios such as images. For CNNs, typically pooling layer defines the operation on a window sliding along images with strides, which shrinks the input size and augments the receptive field of convolution. However, the window and the minified graph are ambiguous due to the diverse topology. Another issue dedicated to our task is that the heterogeneity of brain networks calls for modal-specific designs. Last but not least, practical models should avoid potential computational inefficiency caused by complicated node operation. We prefer graph pooling sharing several appealing properties as CNN:
-
Locality: The pooling windows aggregate local information from neighbor or related nodes.
-
Loyalty: The reduced graph characterizes the structure of primary graph and data.
-
Likely: The computation is efficient. Additionally, the data-driven pooling scheme should be end-to-end trainable, subject to deep learning principle.
To address these challenges, we imitate the process of CNN pooling, and decompose the graph pooling into two steps, node merge and graph distillation [18]. In the first step, nodes are clustered and features are computed accordingly; in the second, graph is reduced for follow-up graph convolution. Formally, the l-th pooling layer is defined as,
here \(H^l\in \mathcal {R}^{n^{l-1}\times k^{l}}\) is the output of graph convolutional layer l, \(P^l\in \mathcal {R}^{n^l\times n^{l-1}}\) is a trainable pooling matrix, \(H_p^l\in \mathcal {R}^{n^{l}\times k^{l}}\) is the output of pooling layer, and \(n^{l}\) and \(k^{l}\) are the number of nodes and feature length, respectively. The reduced graph is defined as,
here \(A^{l}\) is the graph matrix from layer l, and f(A, x) are defined to coincide in the naive regression objective, and \(\Vert \cdot \Vert _F\) is Frobenius norm. The first term can be derived by preserving the graph convolution consistency for pooled nodes,
here \(U_P\) and U are the Fourier basis of after and before pooling, respectively. Naturally, it can be interpreted as the eccentricity of the reduced graph to the graph at hand, and the second term considers the data fidelity. In principle, this pooling structure is defined using \(P^l\) and \(A^l\), learned from both graph and data.
Now we delve into the details and discuss how the aforementioned concerns are resolved through some further consideration. We decompose the pooling matrix,
here \(A^l_p\in \mathcal {R}^{n^l\times n^{l-1}}\) is a sparse assigning matrix, and each row of \(A^l_p\) represents a cluster in the reduced graph. Each cluster aggregates the vicinity of assigned nodes; meanwhile, the co-occurrence of assigned (perhaps distant) nodes represent a high-level relation beyond neighboring. Therefore, locality is attained by sparsity regularization related to \(A^l_p\). Loyalty is also maintained via compelling \(A^l\) to satisfy (2). Combining the above arguments boils down to the objective,
here the first term is the tedious regression loss, \(\lambda _1\) and \(\lambda _2\) are tunable parameters. This formulation only exerts slight computation burden to naive graph convolutional layer in the inference stage; the training is also an end-to-end routine on an integral structure and avoids some potential redundancy [18]. Jointly, these indicate the likely of the adaptive scheme in both training and computation. Finally, each modality is dealt with using individual GCN, with modal-specified graph and pooling setup, which leads to an intuitive explanation for the effectiveness of the proposed method, that heterogeneity is contained in the first stage, modal-fusion in the second.
3 Experiments
3.1 Data Description
We analyzed the data from PPMI (http://www.ppmi-info.org), which includes 145 healthy controls (HC) (mean \(age=66.70\pm 10.95\), 96 males) and 474 subjects with PD (mean age = \(67.33\pm 9.33\), 318 males). No significant differences was identified in age between HC and PD (P = 0.5). We utilized the diffusion-weighted MRI-derived structure connectome to predict several PD clinical scores, including the Montreal Cognitive Assessment (MoCA) Test, the Tremor Dominant (TD) scores and the Postural Instability and Gait Difficulty (PIGD) scores, REM Sleep Behavior Disorder (RBD) scores, the Geriatric Depression Scale (GDS), and the University of Pennsylvania Smell Identification Test (UPSIT).
For each subject’s T1-weighted MRI, we applied ROBEX, a robust automated brain extraction program trained on manually “skull-stripped” MRI data [9], to remove the extra-cerebral tissue. These skull-stripped volumes were carefully examined and manually edited if needed. Anatomical scans were then underwent the standard FreeSurfer (V6.0, http://surfer.nmr.mgh.harvard.edu/) parcellation, based on which 84 cortical and subcortical ROIs are defined.
For each subject’s diffusion-weighted MRI, firstly bet and eddy\(\_\)correct functions in FSL (http://www.fmrib.ox.ac.uk/fsl) were applied to remove the non-brain tissue and correct for the possible distortions, and then the gradient table was adjusted correspondingly for each subject. In order to avoid the distortions at tissue-fluid interfaces, echo-planar induced susceptibility artifacts were corrected by elastically aligning skull-stripped b0 images to each subject’s T1 MRI using Advanced Normalization Tools (ANTs, http://stnava.github.io/ANTs/) with SyN algorithm. The resulted 3D deformation was then applied to the remaining diffusion-weighted volumes to generate the full preprocessed diffusion-weighted MRI data. Finally, based on the 84 ROIs derived from the T1 data, we reconstructed three brain structural graphs using three whole brain probabilistic tractography algorithms, including Orientation Distribution Function-based Hough voting [1] and PICo [14] as well as ball-and-sticks-based Probtrackx [2]. (please refer to [19] for more details). Each brain network was normalized by dividing the maximum values in the matrix to reduce the potential computation biases from the differences in scale and range from different tractography algorithms.
3.2 Experiment Settings
Throughout the experiments, ROIs are defined as vertices, the corresponding rows in each human brain networks are defined as features, and the graph is defined as the average of all brain networks. We compare the proposed method with several related methods: multivariate Ridge Regression (RR), Least Absolute Shrinkage and Selection Operator (LASSO), combined \(l_1\) and \(l_2\) norm (ElasticNet), Neural Networks (NN), and Convolutional Neural Network (CNN). For RR, LASSO and ElasticNet, we search the coefficient of the regularization term ranging from 0.001 to 100 and report the best results. For neural network, we use a two layer structure with 100 hidden units and Relu activation function. For the proposed method, we use two layer GCN for each modality. The feature length for each layer is [16, 32] respectively, and the graph size after pooling is [32, 8]. An one-layer perceptron is used for regression. Both \(\lambda _1\) and \(\lambda _2\) are set to 0.001 in the objective. We use Adam optimizer [10] with a learning rate 0.001, and a batch size of 128. All clinical measures are normalized to [0, 1] by different tests. We reported the root mean square error (RMSE) and mean absolute value (MAE) on 5-fold cross validation as the evaluation metrics.
3.3 Results
We first present the comparison of the performance of the proposed method with multiple baselines, and the results are summarized in Table 1. On both metrics, we observe that the proposed method outperforms baselines consistently. For linear methods, sparse methods achieve better prediction accuracy compared to non-sparse methods, RR. Non-linear models, including NN, CNN, and the proposed model, also improve the prediction performances against linear models in general. The baseline deep methods, NN and CNN, have similar results. Particularly, the proposed method outperforms NN with much less parameters and has better performance with similar parameter size compared to CNN.
In Table 2 we also include the prediction performance of different combination of generated graphs. Obviously, the network using single modal brain network yields much worse prediction, compared to multi-modal networks. The results also imply some interesting observations: particular subsets of involved modals may attain decent results, though the optimal combination require brutal search; the prediction yielded by the network integrating all modals, generally, is the best or at least comparable with the best. To this end it is fair to claim that multi-modality helps the prediction of PD clinical measures, and that more modals are always preferred. Besides, the brain graphs generated by Hough show worth-noting prediction ability, which indicates potential direction for future study.
4 Conclusion
In this paper, we propose a graph convolutional network to predict the PD clinical measures, using multi-modal brain networks. Particularly, we propose an adaptive pooling scheme driven by both graph structure and brain data, which is efficient in computing and end-to-end training. The experiment results demonstrate that the proposed method attains state-of-the-art results compared to related baselines, and integrating multi-modal brain network is highly effective in the prediction task.
References
Aganj, I., et al.: A hough transform global probabilistic approach to multiple-subject diffusion MRI tractography. Med. Image Anal. 15(4), 414–425 (2011)
Behrens, T.E., et al.: Probabilistic diffusion tractography with multiple fibre orientations: what can we gain? Neuroimage 34(1), 144–155 (2007)
Bullmore, E., et al.: Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 10(3), 186 (2009)
Caspell-Garcia, C., et al.: Multiple modality biomarker prediction of cognitive impairment in prospectively followed de novo Parkinson disease. PLoS ONE 12(5), e0175674 (2017)
Defferrard, M., et al.: Convolutional neural networks on graphs with fast localized spectral filtering. In: NeurIPS, pp. 3844–3852 (2016)
Dhillon, I.S., et al.: Weighted graph cuts without eigenvectors a multilevel approach. IEEE TPAMI 29(11), 1944–1957 (2007)
Fornito, A., et al.: Graph analysis of the human connectome: promise, progress, and pitfalls. Neuroimage 80, 426–444 (2013)
Hamilton, W., et al.: Inductive representation learning on large graphs. In: Advances in Neural Information Processing Systems, pp. 1024–1034 (2017)
Iglesias, J.E., et al.: Robust brain extraction across datasets and comparison with publicly available methods. IEEE Trans. Med. Imaging 30(9), 1617–1634 (2011)
Kingma, D.P., et al.: Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980 (2014)
Kipf, T.N., et al.: Semi-supervised classification with graph convolutional networks. arXiv preprint arXiv:1609.02907 (2016)
Ktena, S.I., et al.: Distance metric learning using graph convolutional networks: application to functional brain networks. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017. LNCS, vol. 10433, pp. 469–477. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66182-7_54
Marek, K., et al.: The parkinson progression marker initiative (PPMI). Prog. Neurobiol. 95(4), 629–635 (2011)
Parker, G.J., et al.: A framework for a streamline-based probabilistic index of connectivity (PICO) using a structural interpretation of MRI diffusion measurements. J. Magn. Reson. Imaging Off. J. Int. Soc. Magn. Reson. Med. 18(2), 242–254 (2003)
Shuman, D.I., et al.: The emerging field of signal processing on graphs: extending high-dimensional data analysis to networks and other irregular domains. IEEE Signal Process. Mag. 30(3), 83–98 (2013)
Sporns, O., et al.: The human connectome: a structural description of the human brain. PLoS Comput. Biol. 1(4), e42 (2005)
Xu, K., et al.: How powerful are graph neural networks? arXiv preprint arXiv:1810.00826 (2018)
Ying, Z., et al.: Hierarchical graph representation learning with differentiable pooling. In: Advances in Neural Information Processing Systems, pp. 4805–4815 (2018)
Zhan, L., et al.: Comparison of nine tractography algorithms for detecting abnormal structural brain networks in Alzheimer’s disease. Front. Aging Neurosci. 7, 48 (2015)
Zhang, X., et al.: Multi-view graph convolutional network and its applications on neuroimage analysis for Parkinson’s disease. arXiv preprint arXiv:1805.08801 (2018)
Zhang, Y., Huang, H.: New graph-blind convolutional network for brain connectome data analysis. In: Chung, A.C.S., Gee, J.C., Yushkevich, P.A., Bao, S. (eds.) IPMI 2019. LNCS, vol. 11492, pp. 669–681. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-20351-1_52
Acknowledgements
This study was partially funded by NSF IIS 1836938, DBI 1836866, IIS 1845666, IIS 1852606, IIS 1838627, IIS 1837956, and NIH R21 AG056782, R01 AG049371, U54 EB020403, P41 EB015922, R56 AG058854.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Zhang, Y., Zhan, L., Cai, W., Thompson, P., Huang, H. (2019). Integrating Heterogeneous Brain Networks for Predicting Brain Disease Conditions. In: Shen, D., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2019. MICCAI 2019. Lecture Notes in Computer Science(), vol 11767. Springer, Cham. https://doi.org/10.1007/978-3-030-32251-9_24
Download citation
DOI: https://doi.org/10.1007/978-3-030-32251-9_24
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-32250-2
Online ISBN: 978-3-030-32251-9
eBook Packages: Computer ScienceComputer Science (R0)