Abstract
Effective fusion of multi-modality neuroimaging data, such as structural magnetic resonance imaging (MRI) and fluorodeoxyglucose positron emission tomography (PET), has attracted increasing interest in computer-aided brain disease diagnosis, by providing complementary structural and functional information of the brain to improve diagnostic performance. Although considerable progress has been made, there remain several significant challenges in traditional methods for fusing multi-modality data. First, the fusion of multi-modality data is usually independent of the training of diagnostic models, leading to sub-optimal performance. Second, it is challenging to effectively exploit the complementary information among multiple modalities based on low-level imaging features (e.g., image intensity or tissue volume). To this end, in this paper, we propose a novel Deep Latent Multi-modality Dementia Diagnosis (DLMD\(^2\)) framework based on a deep non-negative matrix factorization (NMF) model. Specifically, we integrate the feature fusion/learning process into the classifier construction step for eliminating the gap between neuroimaging features and disease labels. To exploit the correlations among multi-modality data, we learn latent representations for multi-modality data by sharing the common high-level representations in the last layer of each modality in the deep NMF model. Extensive experimental results on the Alzheimer’s Disease Neuroimaging Initiative (ADNI) dataset validate that our proposed method outperforms several state-of-the-art methods.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
1 Introduction
The Alzheimer’s Disease Neuroimaging Initiative (ADNI) was launched in 2003 by the National Institute on Aging, which collected data from multiple modalities, such as structural magnetic resonance imaging (MRI) [13] and fluorodeoxyglucose positron emission tomography (PET) [2]. The goal of ADNI is to better understand the pathological progression of AD and to identify the most related biomarkers using multi-modality data. Since different modalities provide complementary information, it is critical to effectively fuse multi-modality data to boost diagnostic performance [19, 20].
Recently, several approaches [15] towards multi-view learning(or multi-modality fusion) have been developed and also applied for brain disease diagnosis [6, 18]. As the most straightforward strategy, a simple fusion method is used to pool features from multi-modalities together [5], followed by the training of a classifier (e.g., support vector machine, SVM). However, such a strategy cannot effectively exploit the correlation among multi-modalities, thus leading to sub-optimal diagnostic performance. To effectively fuse multi-modality data, the model in [3] uses Multiple Kernel Learning (MKL) to fuse the data by learning optimal linearly-combined kernels for classification. Additionally, a multi-task learning based feature selection method is proposed in [6], using an inter-modality relationship preserving constraint. Then, Liu et al. [7] uses a zero-masking strategy for data fusion to extract complementary information from multi-modality data. Besides, several multi-view learning methods have been recently proposed for multi-modality fusion, where each modality is treated as a specific view. For example, the Multi-View Dimensionality Co-Reduction (MDCR) method [16] adopts the kernel matching to regularize the dependence across multiple views and projects each view into a low-dimensional space. The Multi-view Learning with Adaptive Neighbours (MLAN) method [8] performs clustering/semi-supervised classification and local structure learning simultaneously. The Deep Matrix Factorization (DMF) method [17] conducts deep semi-nonnegative matrix factorization (NMF) to seek a common representation for the multi-view clustering task. Although considerable progress has been made, there are still several challenges for effective fusion of multi-modality data. First, the fusion of multi-modality data is usually independent of the training of diagnostic models, leading to a sub-optimal performance. Second, it is challenging to effectively exploit the complementary information among multiple modalities based on low-level imaging features.
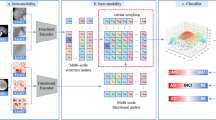
Overview of the proposed DLMD\(^2\) model. It performs deep NMF in a layer-wise manner to learn shared latent representations for multi-modality data, and then projects the new representations into the label space for diagnosis model training. Our method also uses the learned latent representations to reconstruct the original features of multi-modality data, encouraging the new representations to effectively preserve critical and useful information.
To address these issues, we propose a Deep Latent Multi-modality Dementia Diagnosis (DLMD\(^2\)) model to jointly perform high-level feature learning and classifier construction (as shown in Fig. 1). The key idea is to develop a deep NMF model to learn high-level shared latent representations for multi-modality data, whose learned features could have strong interpretability to help uncover the complex structure of the brain. We also reconstruct the original features using the latent representations, making the learned representations to effectively preserve critical and useful information. In addition, the feature learning/fusion of multi-modality data and classification model training are integrated into a unified framework for automated dementia diagnosis. Experimental results on the ADNI dataset show the effectiveness of our DLMD\(^2\) model against other state-of-the-art methods, for several brain disease diagnosis tasks.
In summary, the key contributions of this study are three-fold. (1) A deep NMF model is built using a layer-wise decomposition strategy to effectively uncover the hidden information of multi-modal neuroimaging data. (2) Our model exploits the correlations among multi-modality data by learning shared latent representations for different modalities. (3) Both multi-modality fusion and classification model training are seamlessly integrated into a unified framework for automated dementia diagnosis.
2 Method
Overview of Standard NMF. Consider a non-negative data matrix \(\mathbf {X}=[\mathbf {x}_1,\mathbf {x}_2,\ldots ,\mathbf {x}_n]\in \mathbb {R}^{d\times {n}}\) with n samples, where \(\mathbf {x}_i\,(i=1,\cdots ,n)\) denotes the i-th sample and d is the feature dimension. NMF aims to seek two non-negative matrices \(\mathbf {B}\) and \(\mathbf {H}\), and its objective function is given as
where \(\mathbf {B}\in \mathbb {R}^{d\times {h}}\) denotes the basis matrix, \(\mathbf {H}\in \mathbb {R}^{h\times {n}}\) is regarded as the new representation of the original data \(\mathbf {X}\), and h is the dimension of the new feature representation.
2.1 Proposed Method
Suppose we have a multi-modality neuroimaging dataset \(\{\mathbf {X}^{(1)},\mathbf {X}^{(2)},\ldots ,\mathbf {X}^{(V)}\}\), where \(\mathbf {X}^{(v)}\in \mathbb {R}^{d_v\times {n}}\) denotes the v-th (\(v=1,\cdots ,V\)) modality, \(d_v\) is the dimension of the v-th modality, n is the number of samples, and V is the number of data modalities. The formulation of the multi-modal NMF model can be written as follows
where \(\mathbf {B}^{(v)}\) and \(\mathbf {H}^{(v)}\) denote the basis and representation matrices for the v-th modality, respectively. Using Eq. (2), the new representation can be learned for each modality independently, but the underlying correlation among multiple modalities cannot be captured explicitly. To address this issue, another model can be developed as follows:
where \(\mathbf {H}\) can be considered as the shared representation for different modalities, and can thus be used to exploit the correlation among multiple modalities. In the dementia diagnosis task, we can construct a unified multi-modal feature learning and classifier training framework, defined as
where \(\mathbf {W}\) denotes a projection matrix, and \(\mathbf {Y}\in \mathbb {R}^{c\times {n}}\) is the label matrix with c categories. The model defined in Eq. (4) employs the label information of training data to guide the model to learn discriminative shared representations \(\mathbf {H}\) for multiple modalities. That is, the “good” feature representation learned is expected to boost the classification performance.
Using Eq. (4), we can jointly learn the discriminative shared representation (i.e., \(\mathbf {H}\)) and the classification/diagnosis model. However, one main issue is that Eq. (4) only defines a shallow (i.e., linear) NMF model, which cannot effectively uncover the complex (e.g., high-level) correlations among multiple modalities. It is well known that deep learning can produce high-quality feature representations and also capture the high-level correlations among features. To this end, a deep NMF (or semi-NMF) model has recently been developed [14, 17], with promising results for data representation. Specifically, a multi-layer decomposition process in the deep NMF model is formulated as
where \(\mathbf {B}_l^{(v)}\) (\(l=1,\ldots ,L\)) and \(\mathbf {H}_l^{(v)}\) (\(l=1,\ldots ,L\)) denote the basis matrix and the latent representation matrix of the v-th modality at the l-th layer, respectively. Also, \(\mathbf {H}_L\) is the shared latent representation of different modalities at the last layer, and L is the number of decomposition layers. It is worth noting that the latent representation in the last layer is able to identify shared attributes among different modalities. Thus, the deep NMF model can effectively uncover the correlations among multi-modality data by using the high-level feature representations (i.e., \(\mathbf {H}_L\)).
For an ideal latent representation matrix \(\mathbf {H}_L\), it should be able to reconstruct the original data \(\mathbf {X}^{(v)}\) via the basis matrices with a small reconstruction error, \(\mathbf {X}^{(v)}=\mathbf {B}_{1}^{(v)}\cdots \mathbf {B}_{L}^{(v)}\mathbf {H}_{L}\). On the other hand, it should also be obtained by directly projecting the original data \(\mathbf {X}^{(v)}\) into the latent representation space with the aid of the basis matrix [14], i.e., \(\mathbf {H}_L = {\mathbf {B}_{L}^{(v)}}^{\top }\cdots {\mathbf {B}_{1}^{(v)}}^{\top } \mathbf {X}^{(v)}\). Accordingly, we have the following formulation for each modality as
through which the two components (i.e., the non-negative factorization of the original data \(\mathbf {X}^{(v)}\) and the task-oriented learning of the latent representation \(\mathbf {H}_L\)) guide each other during the learning process. In this way, it is able to obtain the ideal latent representation of the original data.
Finally, we integrate the latent representation learning (via deep NMF) and the classification model construction into a unified framework, and our DLMD\(^2\) model is formulated as follows

where \(\lambda \) and \(\beta \) are trade-off parameters. Besides, \(\mathbf {S}\) is a diagonal matrix used to indicate the labeled samples with \(s_{ii}=1\) if the i-th sample is labeled and 0 otherwise. The label matrix \(\mathbf {Y}=[\mathbf {Y}_{\text {labeled}},\mathbf {Y}_{\text {unlabeled}}]\) includes label information of both labeled and unlabeled subjects, thus ensuring that our model can directly predict labels for unseen test samples.
2.2 Optimization
Initialization. Following [14], we first decompose each modality matrix \(\mathbf {X}^{(v)}\) (i.e., minimize \(\Vert \mathbf {X}^{(v)}-\mathbf {B}_1^{(v)}\mathbf {H}_1^{(v)}\Vert _F^2+\Vert {\mathbf {H}_1^{(v)}}-{\mathbf {B}_1^{(v)}}^{\top }\mathbf {X}^{(v)}\Vert _F^2\)), and then decompose the matrix \(\mathbf {H}_1^{(v)}\) (i.e., minimize \(\Vert \mathbf {H}_1^{(v)}-\mathbf {B}_2^{(v)}\mathbf {H}_2^{(v)}\Vert _F^2+\Vert {\mathbf {H}_2^{(v)}}-{\mathbf {B}_2^{(v)}}^{\top }\mathbf {H}_1^{(v)}\Vert _F^2\)) until all layers are initialized. Note that we initialize \(\mathbf {H}_L\) using \(\mathbf {H}_L={\sum _{v}\mathbf {H}_{L-1}^{(v)}}/{V}\). Then, we utilize an alternative optimization method to optimize the objective function, the detailed steps of which are given as follows.
Step 1: Update \(\mathbf {B}_l^{(v)}\). For the v-th modality, we obtain the following equation for \(\mathbf {B}_l^{(v)}\) by taking the derivative of Eq. (7) w.r.t. \(\mathbf {B}_l^{(v)}\):
where \(\mathrm{\Theta }_{l-1}^{(v)}=\mathbf {B}_1^{(v)}\mathbf {B}_2^{(v)}\cdots \mathbf {B}_{l-1}^{(v)}\), and \(\mathrm{\Omega }_{l+1}^{(v)}=\mathbf {B}_{l+1}^{(v)}\mathbf {B}_{l+2}^{(v)}\cdots \mathbf {B}_L^{(v)}\).
By using the Karush-Kuhn-Tucker (KKT) condition [1], we can derive the following updating rule:
Step 2: Update \(\mathbf {H}_L\). We obtain the following equation for \(\mathbf {H}_L\) by taking the derivative of Eq. (7) w.r.t. \(\mathbf {H}_L\):
where \(\mathrm{\Theta }_{L}^{(v)}=\mathbf {B}_1^{(v)}\mathbf {B}_2^{(v)}\cdots \mathbf {B}_{L}^{(v)}\).
By using the KKT condition, we can obtain the following updating rule:
Step 3: Update \(\mathbf {W}\). The associated optimization problem is given as
Denoting \(\mathbf {I}\) as an identity matrix, we have the following updating rule:
We repeat the above updating rules to iteratively optimize \(\mathbf {B}_l^{(v)}\) (\(l=1,2\ldots ,L;v=1,2,\ldots ,V\)), \(\mathbf {H}_L\), and \(\mathbf {W}\), until the model converges. Our model can find at least a locally optimal solution, by seeking an optimal solution for each convex subproblem alternatively. Additionally, several related works have provided the convergence proof associated with the updating rules in Eqs. (9) and (11) using KKT condition [12]. Therefore, the convergence of our model is easily guaranteed.
3 Experiments
3.1 Materials and Neuroimage Preprocessing
The proposed method was evaluated on 379 subjects with complete MRI and PET data at baseline scan from the ADNI dataset, including 101 Normal Control (NC), 185 Mild Cognitive Impairment (MCI), and 93 AD. Within MCI subjects, we defined progressive MCI (pMCI) subjects as MCI subjects that will progress to AD within 24 months, while sMCI subjects remain stable all the time. Subsequently, there were 71 pMCI and 114 sMCI subjects. The MR images were preprocessed via skull stripping, dura and cerebellum removal, intensity correction, tissue segmentation and template registration. Then the processed MR images were divided into 93 pre-defined Regions-Of-Interest (ROIs), and the gray matter volumes were calculated as MRI-based features. We linearly aligned each PET image (i.e., FDG-PET scans) to its corresponding MRI scan, and the mean intensity value of each ROI was calculated as PET-based features. Table 1 summarizes the demographic information of the subjects used in this study.
3.2 Experimental Settings
We evaluated the effectiveness of the proposed model by conducting three binary classification tasks: MCI vs. NC, MCI vs. AD, and sMCI vs. pMCI classification. We used four popular metrics for performance evaluation, including accuracy (ACC), sensitivity (SEN), specificity (SPE), and Fscore. We compared our method with two conventional methods: (1) Baseline method (Baseline), which concatenates MRI and PET ROI-based features into a vector for an SVM classifier, and (2) MKL method [3]. We further compared our method with four state-of-the-art multi-view/modality learning methods, including (1) shallow NMF [4], (2) MDCR [16], (3) MLAN [8], (4) DMF [17], and Mdl-cw [9]. We performed 10-fold cross-validation with 10 repetitions for all the methods under comparison, and reported the means and standard deviations of the experimental results. For our method, we determined two parameters (i.e., \(\lambda ,\beta \in \{10^{-5},10^{-4},\ldots ,10^2\}\)) and the dimension of each layer (i.e., \(h_l \in \{90,80,\ldots ,20\}\)) via an inner cross-validation search on the training data, and we also set \(L=2\) in Eq. (5). For other methods, we used inner cross-validation to determine hyper-parameter values. Note that our method and MLAN can directly perform disease prediction, while the other methods need to resort to SVM for prediction (the parameter C in SVM is selected from \(\{10^{-5},10^{-4},\ldots ,10^{2}\}\)).
3.3 Results and Discussion
Figure 2 shows the comparison results achieved by seven methods on three classification tasks. Note that five competing methods (i.e., Baseline, MKL, NMF, DMF and MDCR) conduct feature learning and model training via two separate steps, while our model, Mdl-cw and MLAN integrate them into a unified framework. From Fig. 2, it can be clearly seen that our proposed method performs better than all the comparison methods in four metrics. This could be partly because our unified framework ensures the classification model to provide feedbacks to the deep NMF step for focusing on learning discriminative features. Although the DMF method also relies on a deep NMF model, its performance is inferior to ours. One possible reason for this is that DMF only learns the shared representation for multi-modality data, without reconstructing the original features, and it does not use label information to guide the representation learning process (which we do in this work).
Multi-modality Data Fusion. To analyze the benefit of multi-modality fusion, Fig. 3 shows the performance comparison of different methods using multi-modality (i.e., MRI+PET) and single modality (i.e., MRI or PET) data. From Fig. 3, it can be seen that all methods using multi-modality data outperform their counterparts using just a single modality data. However, our method consistently performs better than other comparison methods when using a single modality data (e.g., MRI or PET).
Comparison with State-of-the-Art Methods. We further compare our method with four state-of-the-art methods for pMCI vs. sMCI classification in Table 2. Even though these methods use different numbers of subjects, a rough comparison has demonstrated that our method achieves the best ACC values among the five methods.
4 Conclusion
In this paper, we propose a deep latent multi-modality dementia diagnosis (DLMD\(^2\)) framework, by integrating deep latent representation learning and disease prediction into a unified model. The proposed model is able to uncover hierarchical multi-modal correlations and capture the complex data-to-label relationships. Experimental results on three classification tasks, with both MRI and PET data, clearly validate the superiority of our model over several state-of-the-art methods. Besides, we can extend it to problems with incomplete multi-modality data in the future.
References
Boyd, S., et al.: Convex Optimization. Cambridge University Press, Cambridge (2004)
Chetelat, G., Desgranges, B., De La Sayette, V., Viader, F., Eustache, F., Baron, J.C.: Mild cognitive impairment: can FDG-PET predict who is to rapidly convert to Alzheimer’s disease? Neurology 60(8), 1374–1377 (2003)
Hinrichs, C., Singh, V., Xu, G., Johnson, S.: MKL for robust multi-modality AD classification. In: Yang, G.-Z., Hawkes, D., Rueckert, D., Noble, A., Taylor, C. (eds.) MICCAI 2009. LNCS, vol. 5762, pp. 786–794. Springer, Heidelberg (2009). https://doi.org/10.1007/978-3-642-04271-3_95
Lee, D.D., Seung, H.S.: Algorithms for non-negative matrix factorization. In: NIPS (2001)
Lei, B., Yang, P., Wang, T., Chen, S., Ni, D.: Relational-regularized discriminative sparse learning for Alzheimer’s disease diagnosis. IEEE Trans. Cybern. 47(4), 1102–1113 (2017)
Liu, F., Wee, C.Y., et al.: Inter-modality relationship constrained multi-modality multi-task feature selection for Alzheimer’s disease and mild cognitive impairment identification. NeuroImage 84, 466–475 (2014)
Liu, S., Liu, S., Cai, W., et al.: Multimodal neuroimaging feature learning for multiclass diagnosis of Alzheimer’s disease. IEEE Trans. Biomed. Eng. 62(4), 1132–1140 (2015)
Nie, F., Cai, G., et al.: Auto-weighted multi-view learning for image clustering and semi-supervised classification. IEEE Trans. Image Process. 27(3), 1501–1511 (2018)
Rastegar, S., Soleymani, M., Rabiee, H.R., Mohsen Shojaee, S.: MDL-CW: a multimodal deep learning framework with cross weights. In: CVPR (2016)
Shi, J., Zheng, X., Li, Y., Zhang, Q., Ying, S.: Multimodal neuroimaging feature learning with multimodal stacked deep polynomial networks for diagnosis of Alzheimer’s disease. IEEE J. Biomed. Health Inf. 22(1), 173–183 (2018)
Suk, H.I., Lee, S.W., Shen, D.: Hierarchical feature representation and multimodal fusion with deep learning for AD/MCI diagnosis. NeuroImage 101, 569–582 (2014)
Wang, J., Tian, F., Yu, H., Liu, C.H., Zhan, K., Wang, X.: Diverse non-negative matrix factorization for multiview data representation. IEEE Trans. Cybern. 48(9), 2620–2632 (2018)
Yang, X., Liu, C., Wang, Z., Yang, J., et al.: Co-trained convolutional neural networks for automated detection of prostate cancer in multi-parametric MRI. Med. Image Anal. 42, 212–227 (2017)
Ye, F., Chen, C., Zheng, Z.: Deep autoencoder-like nonnegative matrix factorization for community detection. In: CIKM, pp. 1393–1402. ACM (2018)
Zhang, C., Fu, H., et al.: Generalized latent multi-view subspace clustering. IEEE Trans. Pattern Anal. Mach. Intell. (2018)
Zhang, C., Fu, H., Hu, Q., Zhu, P., Cao, X.: Flexible multi-view dimensionality co-reduction. IEEE Trans. Image Process. 26(2), 648–659 (2017)
Zhao, H., et al.: Multi-view clustering via deep matrix factorization. In: AAAI (2017)
Zhou, T., et al.: Inter-modality dependence induced data recovery for MCI conversion prediction. In: MICCAI (2019)
Zhou, T., Liu, M., Thung, K.H., Shen, D.: Latent representation learning for Alzheimer’s disease diagnosis with incomplete multi-modality neuroimaging and genetic data. IEEE Trans. Med. Imaging (2019)
Zhou, T., Thung, K.H., Zhu, X., Shen, D.: Effective feature learning and fusion of multimodality data using stage-wise deep neural network for dementia diagnosis. Hum. Brain Mapp. 40(3), 1001–1016 (2019)
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Zhou, T. et al. (2019). Deep Multi-modal Latent Representation Learning for Automated Dementia Diagnosis. In: Shen, D., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2019. MICCAI 2019. Lecture Notes in Computer Science(), vol 11767. Springer, Cham. https://doi.org/10.1007/978-3-030-32251-9_69
Download citation
DOI: https://doi.org/10.1007/978-3-030-32251-9_69
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-32250-2
Online ISBN: 978-3-030-32251-9
eBook Packages: Computer ScienceComputer Science (R0)