Abstract
Prediction of treatment responses of hepatocellular carcinoma (HCC) patients, such as local control (LC) and overall survival (OS), from CT images, has been of importance for treatment planning of radiotherapy for HCC. In this paper, we propose a deep learning method to predict LC and OS responses of HCC from abdominal CT images. To improve the prediction efficiency, we constructed a prediction model that learns both the intratumoral information and contextual information between the tumor and the liver. In our model, two convolutional neural networks (CNNs) are trained on each of the tumor image patch and the context image patch, and the features extracted from these two CNNs are combined to train a random forest classifier for predicting the LC and OS responses. In the experiments, we observed that (1) the CNN outperformed the conventional hand-crafted radiomic feature approaches for both the LC and OS prediction tasks, and (2) the contextual information is useful not only individually, but also in combination with the conventional intratumoral information in the proposed model.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Hepatocellular carcinoma (HCC) is the sixth most common cancer and the third most common cause of cancer-related mortality worldwide [1]. Radiation therapy is one of the major options for the treatment of HCC, and the task of predicting the responses of individual patients on radiation therapy is an important process for establishing a treatment plan [2]. The treatment responses include (1) the local control (LC) which determines whether the tumor size is shrinking after the treatment, and (2) the overall survival (OS) which determines the 24-month survival of the patients after the treatment. In terms of binary classification, the LC prediction can be defined as a task of classifying the patient image into one of {LC,PD} classes, where the LC cases show a decrease or increase within 25% in the tumor size, while the progressive disease (PD) cases show tumor size increase of more than 25%. The OS prediction can also be defined as a binary classification of the patient’s survival at a particular period in time, such as 12, 24, or 60 months, after the therapy. Prediction of these types of treatment responses from the patient images before the therapy has been of interest. However, this prediction task is known to be a very challenging problem due to (1) the imaging differences between patients with different responses are ambiguous, and (2) the imaging consistency within the patients with same responses is not high enough to cluster, as shown in Figs. 1 and 2.
A number of works have been suggested to predict the treatment responses of HCC from radiomic feature analysis on the patient images. Tamandl et al. [3] analyzed qualitative measures of CT perfusion, e.g. blood flow, blood volume, and portal liver perfusion, for predicting the early response of hepatocellular carcinoma (HCC). Cozzi et al. [4] performed univariate and multivariate analysis on CT radiomic features including histogram and texture to predict the LC status and overall survival of HCC patients. Zhou et al. [5] predicted the early recurrence of HCC using a logistic regression model with the CT radiomic features of histogram and texture. Shan et al. [6] constructed a prediction model for early recurrence of HCC using CT radiomic features of histogram and wavelet transform. These previous studies have shown that (1) the intratumoral texture is especially useful for predicting the cancer treatment responses, (2) the peritumoral radiomic features extracted from the tumor boundary region showed better prediction performance than the conventional intratumoral features and (3) in addition to the intratumoral information, the contextual information of the region where the tumor interacts with the surrounding organs, provides important information for tumor characteristics. However, most of the previous works have constructed the prediction models with qualitative measures and hand-crafted radiomic features, and deep learning has not yet been applied to predict the treatment response of HCC patients.
In this paper, we propose a deep learning method for predicting the LC and OS treatment responses of HCC patients from abdominal CT images. To reflect both intratumoral and contextual information, two CNN models are trained in parallel on the tumor image patches and the context image patches, respectively. The features extracted from the two trained CNNs are then concatenated to be used for training a random forest classifier to predict the local control response of HCC patients.
2 Methods
Our method consists of three major steps: (1) generation of tumor image patch and context image patch, (2) training of CNNs on two types of patches in parallel, and (3) feature combination and training of random forest classifier. A pipeline of our method is summarized in Fig. 3.
2.1 Generation of Tumor- and Context Image Patches
In CNN-based tumor classification methods, the patch image which is cropped around the tumor has been used for the CNN input instead of an entire image, to mainly reflecting the tumor information. In this patch generation, the information trade-off occurs depending on the ratio of tumor size to the patch size. If most of the patch region is a tumor, the intratumoral texture information can be mainly reflected, but the context information between the tumor and the organ can be ignored. In contrast, if the patch includes not only the tumor but also some part of the organs, the context information would be strengthened, but the reflection of the intratumoral information would be weakened. Thus, we propose a framework to utilize both information by training both types of patches and combining them.
First, the tumor image patch is generated as a squared patch circumscribing the tumor as the conventional image patch for the tumor classifying CNN. The sizes of the tumor image patches with the different sized tumors are resized to match the input size of CNN, i.e. \(227\times 227\) pixels so that the CNN training can be tumor size invariant. The generated tumor image patch can reflect the intratumoral texture and tumor shape information to the CNN training.
Second, the context image patch is generated as a square patch containing both the tumor and the liver. The generated context image patch can reflect the context information between the tumor and the organ, such as the relative location of the tumor in the liver and the organ information itself. Figure 4 shows the examples of the tumor image patches and their corresponding context image patches.
2.2 CNN Training
With the two types of input image patches, we train a CNN on each of these patches to classify the treatment response classes. The AlexNet [7] is used for training, which is one the most used thin-structured CNNs. The AlexNet consists of five convolutional layers, with the first, second, and fifth convolutional layers followed by max-pooling layers. These convolutional parts are then followed by three fully-connected layers, which finally generate the class label probability from the final layer. In our method, the model is pre-trained on the ImageNet database and is fine-tuned on the given dataset. The transfer learning is performed with the learning rate of \(1e^{-4}\), mini-batch size of 20, and maximum epoch of 10.
The trained CNNs can act differently according to the types of input patches. The tumor CNN trained with the tumor image patch mainly learns the correlation between the intratumoral texture information and the treatment response, while the context CNN trained with the context image patch mainly learns the relationship between the context information and the treatment response.
2.3 Feature Combination and Random Forest Training
As a final step, we combine the information of the tumor CNN and the context CNN and perform classifier training on the combined feature to predict the treatment response. In feature combination, we first extract the two 4096-dimensional features from the fc7 layer of both the tumor and the context CNNs. We concatenate the two features to generate a 8192-dimensional combined feature, which contains both intratumoral texture and context information of the tumor.
In the classifier training, the new classifier is trained on the combined features to classify the treatment response classes. In the proposed method, a random forest classifier is used considering the size of a given dataset. In experiments, the number of trees to construct the random forest was experimentally determined to be 100. The data augmentation is also performed on the training set to generate 5,000 images for each class by random rotation, scaling, and translation.
3 Experiments
3.1 Dataset and Experimental Settings
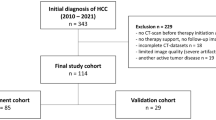
Our study was approved by the local institutional review board (IRB No. 4-2018-0882). Our dataset includes 171 HCC patients with LC and OS treatment response recordings. In terms of LC, the number of patients diagnosed with LC and PD in the dataset was 129 and 42, respectively. In terms of OS, the number of patients diagnosed with Survival and Death in the dataset was 23 and 148, respectively. We used portal phase CT images with the resolution of \(512\times 512\), pixels sizes between \(0.5\times 0.5\) mm2 and \(0.8\times 0.8\) mm2, and slice thicknesses between 3 to 5 mm. From CT scans, all tumors and livers were manually segmented by the clinical expert. For each task, the five-fold cross validation was performed on training and evaluating the classifiers.
To evaluate the performances, we performed the internal and external validation for the proposed method. Pipelines of the comparative methods are summarized in Fig. 5. As an internal validation, the results of the proposed method were compared with those of (1) the tumor CNN only, and (2) the context CNN only. As an external validation, the results of the proposed method were compared with that of random forest classification with the conventional state-of-the-art hand-crafted radiomic features. In this comparative method, the 67-dimensional hand-crafted radiomic features were used including 7 histogram statistics, 5 histogram percentiles, 14 gray-level co-occurrence matrix (GLCM) features, 22 gray-level run-length matrix (GLRLM) features, and local binary pattern (LBP) features, and 9 shape features. A random forest classifier with the same settings as the proposed method was trained on the hand-crafted features. The detailed list of hand-crafted radiomic features used in the comparison method and the details of the comparative method are given in [8]. The performance was evaluated by comparing the accuracy and the area-under-ROC-curve (AUC) for these comparison methods.
3.2 Results for LC Prediction
Table 1 shows the results of LC prediction for the comparative methods. In internal validation, the context CNN showed slightly lower accuracy than the conventional tumor CNN. This might be since the intratumoral texture and shape are known to be significant criteria for predicting the treatment responses as discussed in the previous studies. However, the proposed method combining the two CNN information improved the accuracy by about 5%p and the AUC by about 2%p compared with the tumor CNN. It can be considered that although the context CNN alone may be less accurate than the tumor CNN, but the context information is complementary to the intratumoral information in prediction so that the proposed method combining both information improved overall performances.
In external validation, the conventional hand-crafted radiomic feature classification showed similar performance as the tumor CNN, while the proposed method outperformed it. It can be considered that deep learning has a high potential in the image-based prediction of treatment response for HCC patients.
3.3 Results for OS Prediction
Table 2 shows the results of OS prediction for the comparative methods. It can be observed that the overall performance of OS prediction is higher than those of LC prediction. It can be considered that the image-based learning provides more information in predicting patient survival than predicting tumor growth.
In internal validation, the tumor CNN showed slightly higher accuracy and AUC than the context CNN, as shown in the case of LC prediction. The proposed method combining the two CNN information improved the accuracy by 4.67%p and 5.26%p, compared to the tumor CNN and the context CNN, respectively. It can be considered that the intratumoral information and the context information complement each other for the survival prediction as well as the previous LC prediction, so that the proposed method combining both information improved overall performances.
In external validation, the conventional hand-crafted radiomic feature classification showed similar performance to the context CNN, and showed lower performance than the tumor CNN. The proposed method combining two CNN features improved the performance in accuracy by 5.84%p compared to the hand-crafted feature classification. It can be considered that the CNN can provide more useful information than the conventional radiomic hand-crafted features in not only the LC prediction but also the OS prediction task.
In both LC and OS prediction tasks, the values of AUC are overall distributed at low values. It can be analyzed that the models were biased to the larger sized class, i.e. the LC class for LC prediction and the Death class for OS prediction, due to the class imbalance learning. Further assessment and analysis of the current limitations of the proposed method, and the performance improvement throughout overcoming those problems including the class imbalance remain as future works.
4 Conclusions
In this paper, we proposed a deep learning method to predict LC and OS treatment responses of HCC patients from abdominal CT images. In order to reflect both intratumoral and contextual information, we combined the features extracted from the two CNNs which are trained on the tumor image patch and the context image patch, respectively, and trained a random forest classifier to predict the LC and OS. In experiments, the proposed method achieved higher accuracy compared to not only the existing tumor image-trained CNN, but also the conventional radiomic feature classification. We also verified that the proposed framework can improve the overall performances regardless of prediction target, LC or OS. It can be confirmed that the contextual information is helpful for the prediction of not only the tumor growth but also the survival of HCC patients. Future works include further enhancement of the proposed method throughout the joint learning of CNN and radiomic features and extension of the current classification-based framework to the score regression-level response prediction.
References
Altekruse, S.F., et al.: Hepatocellular carcinoma incidence, mortality, and survival trends in the United States from 1975 to 2005. J. Clin. Oncol. 27, 1485–1491 (2009)
El-Sera, H.B., Mason, A.C.: Rising incidence of hepatocellular carcinoma in the United States. N. Engl. J. Med. 340, 745–750 (1999)
Tamandl, D., et al.: Early response evaluation using CT-perfusion one day after transarterial chemoembolization for HCC predicts treatment response and long-term disease control. Eur. J. Rad. 90, 73–80 (2017)
Cozzi, L., et al.: Radiomics based analysis to predict local control and survival in hepatocellular carcinoma patients treated with volumetric mudulated arc therapy. BMC Cancer 17, 829 (2017)
Zhou, Y., et al.: CT-based radiomics signature: a potential biomarker for preoperative prediction of early recurrence in hepatocellular carcinoma. Abd. Rad. 42, 1695–1704 (2017)
Shan, Q., et al.: CT-based peritumoral radiomics signatures to predict early recurrence in hepatocellular carcinoma after curative tumor resection or ablation. Cancer Imaging 19, 11 (2019)
Krizhevsky, A., et al.: ImageNet classification with deep convolutional neural networks. In: Proceedings of NIPS, pp. 1097–1105 (2012)
Lee, H., et al.: Differentiation of fat-poor angiomyolipoma from clear cell renal cell carcinoma in contrast-enhanced MDCT images using quantitative feature classification. Med. Phys. 44, 3604–3614 (2017)
Acknowledgments
This work was supported by Radiation Technology R&D program through the NRF of Korea (NRF-2017M2A2A7A02070427).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Lee, H., Hong, H., Seong, J., Kim, J.S., Kim, J. (2019). Treatment Response Prediction of Hepatocellular Carcinoma Patients from Abdominal CT Images with Deep Convolutional Neural Networks. In: Rekik, I., Adeli, E., Park, S. (eds) Predictive Intelligence in Medicine. PRIME 2019. Lecture Notes in Computer Science(), vol 11843. Springer, Cham. https://doi.org/10.1007/978-3-030-32281-6_18
Download citation
DOI: https://doi.org/10.1007/978-3-030-32281-6_18
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-32280-9
Online ISBN: 978-3-030-32281-6
eBook Packages: Computer ScienceComputer Science (R0)