Abstract
In this work we tackle the link prediction task in knowledge graphs. Following recent success of Question Answering systems in outperforming humans, we employ the developed tools to identify and verify new links. To identify the gaps in a knowledge graph, we use the existing techniques and combine them with Question Answering tools to extract concealed knowledge. We outline the overall procedure and discuss preliminary results.
This work has been partially funded by the project LYNX. The project LYNX has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement no. 780602. More information is available online at http://www.lynx-project.eu.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Knowledge graphs (KGs) contain knowledge about the world and provide a structured representation of this knowledge. Current KGs contain only a small subset of what is true in the world [7]. There are different types of information that could be incomplete, for example, incomplete set of entities (not all movies are mentioned in the movies KG), incomplete set of predicates, or missing links between existing entities. Different types of incompleteness are usually addressed with different methods. For example, Named Entity Recognition is successfully applied to find new entities of given classes [10]. In this work we consider the latter problem of link prediction (LP), i.e. finding triples (subject s, predicate p, objects o), where p is defined in the schema of the KG and s and o are known instance contained in the existing KG.
LP approaches can roughly be subdivided into two classes:
-
Leveraging the knowledge from the existing KG
-
The rule induction methods [3, 6] learn rules over KG that capture patterns in data. In a generic domain one can learn, for example, that a person has a home address or that a consumer good has a price. These rules help to identify potential gaps in an incomplete KG. In order to fill in the gaps one needs to verify the veracity of the potential new triple.
-
Embeddings project symbolic entities and relations into continuous vector space. The vector arithmetic is used to predict new links [8, 12].
-
-
Extracting the knowledge from other sources.
-
Transformation of the information from some structured sourceFootnote 1.
-
Relation Extraction (RE) methods [11] employ trained models to recognize triples in the text and add those triples to the existing KG.
-
In this work we consider combining both approaches. Given a KG we use rules and heuristics to identify potential gaps in the KG. Then we employ a Question Answering (QA) framework to find and/or verify new triples. Therefore, for each new triple we can demonstrate supporting piece of text. It is interesting to compare with the paper [1] describing a ranking method to provide descriptive explanations for relationships in the KG. In our paper we look for relationships expressed in the text that are not yet in the KG.
The difference of the introduced methodology from the existing approaches:
-
LP typically considers only the KG itself and not the textual data.
-
RE lacks in finding new relations not presented in the training set.
2 Approach
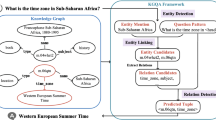
After applying the learned rules or heuristics we obtain a set of pairs (s, p) and triples (s, p, o). In case of having pairs, the task is to find objects O such that \(\{(s,p,o)\ |\ o \in O\}\) is a set of valid triples. In case of having triples, the task is to verify the provided triples. Our approach consists of the following steps:
-
1.
Question formulation,
-
2.
Retrieving documents potentially containing answers from the corpus,
-
3.
Employing QA over documents to get candidate answers and their scores,
-
4.
Choosing correct answers.
Question Formulation. The goal is to go from a pair (s, p) to such a question that a correct answer o defines a valid triple (s, p, o). In order to articulate this natural language question q we employ lexicalization techniques [5].
Retrieving Documents. Given a question q we use query expansion techniques [2] and formulate a search query to retrieve relevant document from the corpus.
Question Answering. Our end-to-end QA system accepts a natural language question and a set of documents as input and outputs a set of pairs (answer, confidence score). The score indicates the confidence of the system that the answer is correct. The QA systems are usually computationally demanding, therefore it is not feasible to send the whole corpus as an input to the QA system.
We use an implementation of BERT [4] that we trained on a large set of question-answer pairs of SQuAD dataset [9]. BERT is a deep learning based system that outperforms previous approaches and reaches close to human performance in QA tasks.
Verification and Integration. Given the answers produced by the QA system we try to match each answer to an instances in the KG. If an answer could not be matched then it is discarded. For the undiscarded answers we check if the class of the matched instances complies with the range restrictions of the predicate definition in the schema. If the range restriction is satisfied we add the triple (subject, predicate, matched instances) to the KG.
In the special case when we know that at most one triple is allowed for a pair (subject, predicate) we choose the answer yielding the highest score of the QA system if it satisfies the range restriction.
3 Preliminary Results and Conclusion
The first used corpus is a collection of over 3300 biomedical paper abstracts from “PubMed”Footnote 2 containing term “BRCA1”. Questions and first 3 answers are provided in the Table 1. For the first question: “Which diseases does niraparib treat?” it is possible to check the following triple (niraparib, treatsDisease, BRCA-mutant ovarian cancer). If this triple fails the verification stage, we continue to verify other candidates for their existence. In this case the answer satisfies the verification as “BRCA-mutant ovarian cancer” is indeed a disease.
In all three examples we asked what kind of disease is treated by such drugs as niraparib, rucaparib, and olaparib. All these drugs are anti-cancer agents, the system can successfully match these drugs with treating cancer. Moreover, the system is able to identify the specific types of cancer and additional details.
The second dataset is a collection of paper abstracts containing the term “rs1045642”, the identifier of a single-nucleotide polymorphism (SNP) in the human genome. We intend to observe data about mutations, the KG of which we expect not to be complete as the database of mutations is not up-to-date. To add such frequently updated data one should inspect corresponding literature. That being said, it is clear that an automated system could come as a benefit.
Conclusion. We considered an important practically relevant task of LP in KGs. In our approach we combine existing techniques to identify potential gaps with QA system to extract concealed knowledge from a text corpus and to formulate new triples. The first experiments show promising results even for domain specific datasets.
References
Bhatia, S., Dwivedi, P., Kaur, A.: That’s interesting, tell me more! Finding descriptive support passages for knowledge graph relationships. In: Vrandečić, D., et al. (eds.) ISWC 2018. LNCS, vol. 11136, pp. 250–267. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00671-6_15
Bhogal, J., Macfarlane, A., Smith, P.: A review of ontology based query expansion. Inform. Process. Manage. 43(4), 866–886 (2007)
d’Amato, C., Staab, S., Tettamanzi, A.G.B., Minh, T.D., Gandon, F.: Ontology enrichment by discovering multi-relational association rules from ontological knowledge bases. In: ACM, vol. 31, pp. 333–338 (2016)
Devlin, J., Chang, M., Lee, K., Toutanova, K.: BERT: pre-training of deep bidirectional transformers for language understanding. CoRR abs/1810.04805 (2018)
Ell, B., Harth, A.: A language-independent method for the extraction of RDF verbalization templates. In: INLG 2014, pp. 26–34 (2014)
Ho, V.T., Stepanova, D., Gad-Elrab, M.H., Kharlamov, E., Weikum, G.: Rule learning from knowledge graphs guided by embedding models. In: Vrandečić, D., et al. (eds.) ISWC 2018. LNCS, vol. 11136, pp. 72–90. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00671-6_5
Ji, G., He, S., Xu, L., Liu, K., Zhao, J.: Knowledge graph embedding via dynamic mapping matrix. In: ACL 2015, Volume 1: Long Papers, pp. 687–696 (2015)
Lin, Y., Liu, Z., Sun, M., Liu, Y., Zhu, X.: Learning entity and relation embeddings for knowledge graph completion. In: Proceedings of the Twenty-Ninth AAAI Conference on Artificial Intelligence, pp. 2181–2187 (2015)
Rajpurkar, P., Jia, R., Liang, P.: Know what you don’t know: unanswerable questions for squad. In: Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics, pp. 784–789 (2018)
Sanchez-Cisneros, D., Aparicio Gali, F.: UEM-UC3M: an ontology-based named entity recognition system for biomedical texts. In: SemEval 2013, pp. 622–627. Association for Computational Linguistics (2013)
Schutz, A., Buitelaar, P.: RelExt: a tool for relation extraction from text in ontology extension. In: Gil, Y., Motta, E., Benjamins, V.R., Musen, M.A. (eds.) ISWC 2005. LNCS, vol. 3729, pp. 593–606. Springer, Heidelberg (2005). https://doi.org/10.1007/11574620_43
Wang, Z., Zhang, J., Feng, J., Chen, Z.: Knowledge graph embedding by translating on hyperplanes. In: AAAI 2014, pp. 1112–1119 (2014)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Khvalchik, M., Revenko, A., Blaschke, C. (2019). Question Answering for Link Prediction and Verification. In: Hitzler, P., et al. The Semantic Web: ESWC 2019 Satellite Events. ESWC 2019. Lecture Notes in Computer Science(), vol 11762. Springer, Cham. https://doi.org/10.1007/978-3-030-32327-1_23
Download citation
DOI: https://doi.org/10.1007/978-3-030-32327-1_23
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-32326-4
Online ISBN: 978-3-030-32327-1
eBook Packages: Computer ScienceComputer Science (R0)