Abstract
Alzheimer’s Disease (AD) is the main cause for age-related dementia. Many machine learning methods have been proposed to identify important genetic bases which are associated to phenotypes indicating the progress of AD. However, the biological knowledge is seldom considered in spite of the success of previous research. Built upon neuroimaging high-throughput phenotyping techniques, a biological knowledge guided deep network is proposed in this paper, to study the genotype-phenotype associations. We organized the Single Nucleotide Polymorphisms (SNPs) according to linkage disequilibrium (LD) blocks, and designed a group 1-D convolutional layer assembling both local and global convolution operations, to process the structural features. The entire neural network is a cascade of group 1-D convolutional layer, 2-D sliding convolutional layer and a multi-layer perceptron. The experimental results on the Alzheimer’s Disease Neuroimaging Initiative (ADNI) data show that the proposed method outperforms related methods. A set of biologically meaningful LD groups is also identified for phenotype discovery, which is potentially helpful for disease diagnosis and drug design.
This study was partially supported by U.S. NSF IIS 1836945, IIS 1836938, DBI 1836866, IIS 1845666, IIS 1852606, IIS 1838627, IIS 1837956, and NIH AG049371. NIH AG056782.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
1 Introduction
Alzheimer’s disease (AD) is a chronic neuro-degenerative disease. It is the 5th leading cause of death for those aged 65 or older [2] in the United States, and 5.5 million people have Alzheimer’s dementia by estimation. In 2017, total payments are estimated at \(\$259\) billions for all individuals with dementia.
Based on neuroimaging and genetics techniques, computational methods [4, 14] have been shown as powerful tools for understanding and assistant diagnosis on AD. As a result, many SNPs identified as risk factors for AD are listed by the AlzGene database (www.alzgene.org). Among these, early efforts attempting on discovering the phenotype-genotype association mainly exploit linear regression models. Classical methods [1, 10] apply individual regression at all time points, ignoring potential progressive variations of brain phenotypes. Based on linear models, sparse learning [14, 21] is demonstrated to be highly effective. The Schatten p-norm is used to identify the interrelation structures existing in the low-rank subspace [16], with the idea further developed by capped trace norm [5]. Regularized modal regression model [15] is found to be robust to outliers, heavy-tailed noise, and skewed noise. Clustering analysis [6], particularly auto-learning based methods [17] to extract group structures can further help uncover the interrelations for multi-task regression.
However, the aforementioned methods are driven mainly by machine learning principles. Given the complex relation of significant SNP loci identification, an emerging interest is to integrate advanced models and biological priors simultaneously for the phenotype-genotype prediction task. In this paper, we propose a biological knowledge guided convolution neural network (CNN) benefiting from both principles of sparse learning and non-linearity, to address the genotype-phenotype prediction problem. SNPs are structural features equipped with biological priors, and in this paper, we group SNPs according to the Linkage Disequilibrium (LD) blocks, which refers to alleles that do not occur randomly with respect to each other in two or more loci. Based on LD blocks, we develop a group 1-D convolutional layer. Our design modifies naive convolutional layer by applying both group and global convolution, capable of handling structural features as well as utilizing the sparsity of SNP-LD patterns. The entire structure is composed of a cascade of the novel group 1-D convolutional layer, 2-D sliding convolutional layer, and multi-layer perceptron. The experiments verify the impression that sparse learning, group structure, and non-linearity indeed provide improvements, and the proposed method outperforms the related baselines. Also, some interesting LD blocks are identified as important biomarkers for predicting phenotypes.
The rest of this paper is organized as follow: Sect. 2 describes the biological knowledge guided neural network; Sect. 3 shows the experiment results on the Alzheimer’s Disease Neuroimaging Initiative (ADNI) data; Sect. 4 concludes the paper.
2 Methodology
2.1 Problem Definition and Methodology Overview
The proposed method aims at the genotype-phenotype prediction. The SNPs of subject i are features with group structures \(X_i= [g_{i,1},g_{i,2},\ldots ,g_{i,c}]\), where \(g_{i,j}\in \mathbb {R}^{c_j}\) is a feature vector of group j with size \(c_j\), and c is the number of groups. In the proposed method, the SNPs are grouped according to the LD blocks. The phenotypes of subject i are denoted as vector \(Y_i\in \mathbb {R}^m\), where m is the number of phenotypes. The task is to learn a model capturing the association between \(X_i\) and \(Y_i\).
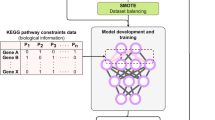
In this paper, we design a biological knowledge guided deep neural network based on SNP feature structures. As illustrated in Fig. 1, the network takes sparse group feature \(X_i\) as the inputs and yields the phenotype prediction \(\hat{Y}_i\) as the outputs. The pipeline of the proposed method involves three steps: first, we obtain the embeddings encoding both local and global information of the structural SNP features, via the novel group 1-D convolutional layer; then, the embeddings are fed into the 2-D sliding convolutional layer for a secondary feature extraction; and at last, the prediction of phenotypes are attained using a multi-layer perceptron.
2.2 Biological Knowledge Guided CNN for Genotype-Phenotype Prediction
Naive convolution layer is not an immediate solution to the phenotype-genotype prediction problems, and the reason is two-folded. First, by definition naive convolution layer is a global operation, thus fails to exploit the group structure of SNPs. Second, the embeddings from the global operation extract the entangled relations across SNPs, blurring the association within specific LD blocks and associated phenotypes. In order to overcome these difficulty, we propose to use additional local convolutional operations on each LD block, besides the global convolutional operation encoding all SNPs. The embeddings yielded by both local and global convolution are then concatenated for the successive 2-D convolution. Formally, the group 1-D convolutional layer takes structural features \(X_i\) as input and yields concatenated embeddings \(h_i\). A local 1-D convolution and max-pooling is operated on each group. For group j, the feature is \(g_{i,j}\in \mathbb {R}^{c_j}\), and the 1-D convolution kernel is \(f_j\in \mathbb {R}^{k_j\times d_h}\), here \(k_j\) is the kernel size for 1-D convolution, \(d_h\) is the number of channels. The output of a local convolution is thus,
here \(*\) is the convolution operation channel-wise, \(P(\cdot )\) is 1-D max-pooling operation along each channel, and \(h_{i,j}\) is the output of the local convolutions and pooling. The output \(h_{i,j}\in \mathbb {R}^{d_h}\). Besides group-wise local operations, a global convolution is also applied. The global convolution is similarly defined as Eq. (2), by replacing the input with concatenated group features. The embeddings of all local convolutions and the global convolutions are then concatenated, \(h_i = [h_{i,1},h_{i,2},\ldots ,h_{i,c},h_{i,0}]\), here \(h_{i,0}\) refers to the output of the global 1-D convolution, \(h_i\in \mathbb {R}^{(c+1)\times d_h}\) is the layer output defined above, and c is the number of groups. Throughout this layer, each group has a distinct convolutional kernel. The kernel size can be also tuned group-wise, as long as the channels are identical to keep the consistency of concatenation. In our data, the SNPs belongs exclusively to each LD blocks, and the size of local convolution is quite limited. Therefore we choose a shared-size local 1-D convolution for all groups, and a separate large-size 1-D convolution kernel for the global embeddings.
The embeddings from group 1-D convolutions layer can be viewed as “sentence” describing the genetic variations. Inspired by TextCNN [8], we design a variant of the standard convolutions layer. Different from images where the spatial coordinates are isotropic, in the embeddings of group SNPs, the convolution kernels are supposed to take complete block information into consideration. Thus we define the receptive field on entire neighboring block. Formally, we define the 2-D convolution kernel \(f^c\in \mathbb {R}^{d_h\times d_p \times k_c}\), here the \(k_c\) is the number of channels, \(d_h\times d_p\) is the kernel size, and \(d_h\) is the size of the concatenated embeddings defined above. Max-pooling \(P(\cdot )\) is utilized after the convolution operations. We use \(h_{i}\) to denote the output of the sparse group layer and \(h_{i}^{2d}\) to denote the output of the 2-D sliding convolutional layer,
the input to the 2-D convolution layer is not padded, thus and the dimension of output of one channel after the convolution operation is \(c+2-k_c\). \(P(\cdot )\) is max-pooling operation along each channel as in group 1-D convolution layer, and the output of 2-D convolution layer \(h_{i}^{2d}\in \mathbb {R}^{k_c}\), which is a vector ready for multi-layer perceptron. We adopt a standard MLP and mean squared error objective for regression.
The proposed 1-D group layer can also be interpreted as a method with pre-defined sparsity, as that the response of a local convolution can be obtained by padding zeros on group features to full size, and discarding the zero responses of padded areas after the convolution operation. Compared to classical methods such as LASSO and group LASSO, the proposed method avoids some drawbacks. In standard LASSO method, frequently only one feature is selected arbitrarily from a set of highly correlated SNPs, particularly those grouped by LD blocks, potentially misleading the identification of important SNPs. Via the proposed group convolution, the contributions of LD blocks are properly assigned to each SNPs involved. On the other, group LASSO method applies group selections, which potentially overlooks the cross-group contributions of SNPs. This phenomenon can be alleviated by the global convolution operations.
Though the coincidence of the terminology, the proposed method is fundamentally different from previous methods exploiting the idea of processing channels of convolutional layers by groups, such as ShuffleNet [22] or ResNeXt [20]. Previous methods typically define their kernels on the entire inputs, and group the middle layer embeddings to improve computational efficiency. These networks are designed to handle large-scale problems, and the convolution kernels are still global. However in the proposed group 1-D convolutional layer, the groups are a pre-defined biological structures on SNPs, thus only proper parts of the features are visible for corresponding local convolutions operations. To sum up, the “group” in the proposed method is used under a different meaning with previous methods.
3 Experiments
3.1 Data Description
The experiments are conducted on the data obtained from the Alzheimer’s Disease Neuroimaging Initiative(ADNI) database (adni.loni.usc.edu). In our experiment the genotype data [11] included all non-Hispanic Caucasian participants from the ADNI Phase 1 cohort which were genotyped using the Human 610-Quad BeadChip. Pre-processing on the SNP data include the standard quality control (QC) and imputation. The QC criteria are composed of (1) call rate check per subject and per SNP marker, (2) gender check, (3) sibling pair identification, (4) the Hardy-Weinberg equilibrium test, (5) marker removal by the minor allele frequency and (6) population stratification. The QC’ed SNPs were then imputed using the MaCH software [9] to estimate the missing genotypes. Among all, we selected only SNPs within the boundary of \(\pm 20K\) base pairs of the 153 AD candidate genes, listed on the AlzGene database (www.alzgene.org) as of 4/18/2011 [3]. As a result, our analyses included 3,576 SNPs extracted from 153 genes (boundary: \(\pm 20K\)) using the ANNOVAR annotation (http://www.openbioinformatics.org/annovar/). The groups of SNPs are defined using the linkage disequilibrium (LD) blocks, and a total of 800 blocks are marked. 2098 SNPs are selected with sufficient group information.
Each MRI T1-weighted image was anterior commissure posterior commissure (AC-PC) aligned using MIPAV2, intensity in-homogeneity corrected using the N3 algorithm [13], skull stripped with manual editing [19], and cerebellum-removed [18]. The image is segmented using FAST [23] into gray matter (GM), white matter (WM), and cerebrospinal fluid (CSF), and registered to a common space using HAMMER [12]. GM volumes normalized by the total intracranial volume were extracted as features, from 93 ROIs defined in [7]. The data for the experiments includes all 737 subjects with sufficient data.
3.2 Experiment Settings
We compare the proposed method with several other regression methods: multivariate Linear Regression(LR), multivariate Ridge Regression(RR), Least Absolute Shrinkage and Selection Operator(LASSO), the combination of L1 norm ad L2 norm (Elastic) [14], Temporal Structure Auto-Learning Predictive Model (TSALPM) [17], Regularized Modal Regression (RMR) [15], and naive CNN. For both RR and LASSO, we set the coefficient of the regularization term through grid search ranging from 0.01 to 10. For TSALPM and RMR, we use the default parameter settings in original papers. For naive CNN, we replace the group 1-D convolutional layer with naive 1-D convolutional layer, and keep the output channels consistent with the proposed method for a fair comparison. For the proposed method, we used one sparse local CNN layer, one CNN layer and two-layer perceptron. We used a shared-size local CNN for each group, with \(k_j = 10\) and \(d_h = 5\). For global 1-D CNN, the kernel size is chosen as \(k_j = 100\). The kernel of the first CNN layer is 400. The hidden units of the two-layer perceptron is 200, and we use a dropout rate of 0.5 during training. The coefficient of regularization term in the objective, \(\lambda \), is 0.01. We use the momentum stochastic gradient descent optimization method with a learning rate of 0.001, and the momentum coefficient of 0.8. The batch size during training is 50. The scores of each ROIs are normalized to zero mean and unit variance. We reported the average root mean square error (RMSE) and mean absolute value (MAE) for each method on five runs.
3.3 Results and Discussions
We compare the proposed method against related baselines and the results are summarized in Table 1. On both metrics, we observe that the proposed method outperforms baselines consistently. For linear methods, sparse methods including LASSO, Elastic, and TSALPM achieve better prediction accuracy compared to non-sparse methods, LR and RR. Non-linear models, including RMR, naive CNN, and the proposed model, also improve the prediction performances against linear models in general. The group information exploited by the proposed sparse group CNN further improves the performance compared to all baselines on both metrics. The results of the proposed method shows decent improvements over naive CNN without sparse group layer. Naive CNN overlooks the group information. Meanwhile, though the dropout layer during training exerts potentially implicit sparsity in naive CNN, the linkage disequilibrium blocks in SNPs still provides additional sparse structure given our task. This demonstrates that the biological knowledge group structure is highly effective in the genotype-phenotype prediction problem.
We also include the top-selected linkage disequilibrium blocks with respect to phenotypes. The importance for LD blocks are defined as \(\mathbb {I}_c = \sum _{i\in c,k,j}{|g_{i,j}^{k}|}\), here \(\mathbb {I}_c\) is the importance of linkage disequilibrium block c, \(i\in c\) is SNP belonging to c, and \(g_{i,j}^{k}\) is the gradient accordingly. The results are summarized in Fig. 2. Some most notable LD blocks from our experiments are: (1) group 629, includes OLR1 1050289, OLR1 10505755, OLR1 3736234 and OLR1 3736233; (2) group 349, includes CTNNA3 1948946 and CTNNA3 13376837; (3) group 363, includes CTNNA3 10509276, CTNNA3 713250, CTNNA3 12355282. We expect the results here indicate some complex interactions within these LD blocks, which is promising for experimental assessment.
4 Conclusion
In this paper we propose a biological knowledge guided convolutional neural network to address the genotype-phenotype prediction problem. We use LD blocks to group SNPs, and develop a novel group 1-D convolutional layer to process the structural features. The prediction is attained through the sequential network of group 1-D convolutional layer, 2-D sliding convolutional layer, and a multi-layer perceptron. The experiments on ADNI data show that the proposed method outperforms related methods. Particularly, the experiments demonstrate the effectiveness of sparse structure compared to dense methods, and the advantage of local CNN layer against naive CNN methods. We also identify a set of biologically meaningful LD blocks for biomarker discovery, which is potentially helpful for disease diagnosis and drug design.
References
Ashford, J.W., Schmitt, F.A.: Modeling the time-course of alzheimer dementia. Curr. Psychiatry Rep. 3(1), 20–28 (2001)
Association, A., et al.: 2017 alzheimer’s disease facts and figures. Alzheimer’s Dement. 13(4), 325–373 (2017)
Bertram, L., McQueen, M.B., Mullin, K., Blacker, D., Tanzi, R.E.: Systematic meta-analyses of alzheimer disease genetic association studies: the alzgene database. Nat. Genet. 39(1), 17 (2007)
Brun, C.C., et al.: Mapping the regional influence of genetics on brain structure variability–a tensor-based morphometry study. Neuroimage 48(1), 37–49 (2009)
Huo, Z., Shen, D., Huang, H.: Genotype-phenotype association study via new multi-task learning model. Pac. Symp. Biocomput. World Sci. 23, 353–364 (2017)
Jin, Y., et al.: Automatic clustering of white matter fibers in brain diffusion MRI with an application to genetics. Neuroimage 100, 75–90 (2014)
Kabani, N.J., MacDonald, D.J., Holmes, C.J., Evans, A.C.: 3D anatomical atlas of the human brain. Neuroimage 7(4), S717 (1998)
Kim, Y.: Convolutional neural networks for sentence classification. arXiv preprint (2014). arXiv:1408.5882
Li, Y., Willer, C.J., Ding, J., Scheet, P., Abecasis, G.R.: Mach: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 34(8), 816–834 (2010)
Sabatti, C., et al.: Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat. Genet. 41(1), 35 (2009)
Saykin, A.J., et al.: Alzheimer’s disease neuroimaging initiative biomarkers as quantitative phenotypes: genetics core aims, progress, and plans. Alzheimer’s Dement. 6(3), 265–273 (2010)
Shen, D., Davatzikos, C.: Hammer: hierarchical attribute matching mechanism for elastic registration. In: Proceedings IEEE Workshop on Mathematical Methods in Biomedical Image Analysis (MMBIA 2001), pp. 29–36. IEEE (2001)
Sled, J.G., Zijdenbos, A.P., Evans, A.C.: A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans. Med. imag. 17(1), 87–97 (1998)
Wang, H., et al.: From phenotype to genotype: an association study of longitudinal phenotypic markers to alzheimer’s disease relevant snps. Bioinformatics 28(18), i619–i625 (2012)
Wang, X., Chen, H., Cai, W., Shen, D., Huang, H.: Regularized modal regression with applications in cognitive impairment prediction. In: Advances in Neural Information Processing Systems, pp. 1448–1458 (2017)
Wang, X., Shen, D., Huang, H.: Prediction of memory impairment with MRI data: a longitudinal study of alzheimer’s disease. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9900, pp. 273–281. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46720-7_32
Wang, X., et al.: Longitudinal genotype-phenotype association study via temporal structure auto-learning predictive model. In: Sahinalp, S.C. (ed.) RECOMB 2017. LNCS, vol. 10229, pp. 287–302. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-56970-3_18
Wang, Y., et al.: Knowledge-guided robust mri brain extraction for diverse large-scale neuroimaging studies on humans and non-human primates. PloS One 9(1), e77810 (2014)
Wang, Y., Nie, J., Yap, P.-T., Shi, F., Guo, L., Shen, D.: Robust deformable-surface-based skull-stripping for large-scale studies. In: Fichtinger, G., Martel, A., Peters, T. (eds.) MICCAI 2011. LNCS, vol. 6893, pp. 635–642. Springer, Heidelberg (2011). https://doi.org/10.1007/978-3-642-23626-6_78
Xie, S., Girshick, R., Dollár, P., Tu, Z., He, K.: Aggregated residual transformations for deep neural networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1492–1500 (2017)
Yang, T., et al.: Detecting genetic risk factors for alzheimer’s disease in whole genome sequence data via lasso screening. In: 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), pp. 985–989. IEEE (2015)
Zhang, X., Zhou, X., Lin, M., Sun, J.: Shufflenet: an extremely efficient convolutional neural network for mobile devices. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 6848–6856 (2018)
Zhang, Y., Brady, M., Smith, S.: Segmentation of brain mr images through a hidden markov random field model and the expectation-maximization algorithm. IEEE Trans. Med. Imag. 20(1), 45–57 (2001)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Zhang, Y., Zhan, L., Thompson, P.M., Huang, H. (2019). Biological Knowledge Guided Deep Neural Network for Brain Genotype-Phenotype Association Study. In: Zhu, D., et al. Multimodal Brain Image Analysis and Mathematical Foundations of Computational Anatomy. MBIA MFCA 2019 2019. Lecture Notes in Computer Science(), vol 11846. Springer, Cham. https://doi.org/10.1007/978-3-030-33226-6_10
Download citation
DOI: https://doi.org/10.1007/978-3-030-33226-6_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-33225-9
Online ISBN: 978-3-030-33226-6
eBook Packages: Computer ScienceComputer Science (R0)