Abstract
Diabetic retinopathy (DR) is a diabetes complication that affects retina due to diabetes mellitus. Funduscopy capture the phenomena, however, it needs to be augmented by effective algorithms that enable detection of severity levels of DR from fundus images. Researchers have mostly proposed conventional convolutional neural networks (CNNs) for binary classification with a very few attempts on multi-class detection of DR. In this paper, unlike other approaches, we propose a deep All-CNN architecture for the detection of DR and its five levels. We first correct the fundus images against the sensor parameters by using Gaussian filters and perform blending in order to highlight foreground features. This is followed by removal of retinal boundaries that further helps the detection process. The proposed pre-processing helps in visualization of intrinsic features of the images as well as builds trust in predictions using the model. Thus pre-processed images are fed into the proposed All-CNN architecture. This has 10 convolutional layers and a softmax layer for the final classification. It includes three convolutional layers with kernel size \(3\times 3\) at strides of 2 which are designed to work as pooling layers; while two convolutional layers with kernel size \(1\times 1\) at strides of 1 are constructed to act as fully connected layers. We apply the proposed methodology on the publicly available Kaggle dataset that has the five-class labeled information. Our model is trained on 30000 retinal fundus images while tested on 3000 images. The proposed architecture is able to achieve 86.64% accuracy, loss of 0.46, and average F1 score of 0.6318 for the five classes. Unlike other architectures, our approach: (1) is All-CNN architecture, (2) provides multi-class information of DR, and (3) has outperformed existing approaches in terms of accuracy, loss and F1 score.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Worldwide diabetic patients are about 280 millions which are estimated to be doubled by 2025 [1]. It is predicted that more than 30% of the diabetic patients will be within the Asia Pacific region by the end of 2025 [2]. In particular, according to the World Health Organization (WHO), 31.7 million people were affected by diabetes mellitus (DM) in India in the year 2000 and this figure is estimated to rise upto 79.4 million by 2030, largest across the world [3].
One of the serious drawbacks of diabetes is an eye disease called diabetic retinopathy (DR) that damages retina leading to the blindness. It is estimated that globally around 15000 to 39000 people loose their sights because of the DR. In the world, around 14.6% aged 40 years and above suffered with the DR after a 5-year duration of diabetes [4]. Note that the DR affects upto 80% of those who had diabetes for 20 years or more [5]. Hence this urge for prevention, early detection, and diagnosis of DR as well as its severity scales.
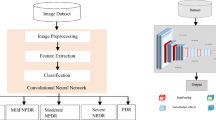
In this paper, we focus on developing algorithmic approach for multi-class classification of the DR from retinal fundus images. Note that the researchers have mostly worked on developing algorithms for DR or No-DR [6,7,8]. However, according to the DR severity scale, it is divided into five classes: No DR, mild non proliferative DR (NPDR), moderate NPDR, severe NPDR, and proliferative DR (PDR). In particular, features like microaneurysms, hemorrhages and exudates are observed in retina during the NPDR stage. In PDR stage, abnormal blood vessels are formed at the back of eye called neovascularization that bursts and bleeds, and finally leads to the blur vision. Hence it is important to detect the stages of DR.
In the recent years, deep convolutional neural network (CNN) has shown excellent performance in information processing for medical data [9,10,11]. One among the CNNs is All-CNN, where pooling and fully connected layers are replaced by the convolutional layers. Experimental results in [12] has shown that inspite of its simplicity, All-CNN performs better than the conventional CNN with a minimum error rate for CIFAR-10 dataset. In this paper, we propose a deep learning based All-CNN architecture for the multi-class classification of diabetic retinopathy on publicly available Kaggle dataset [13].
2 Related Work
Recently classification of diabetic retinopathy becomes an active area of research. In [14], images are augmented by rotation and translation for class balance, and pre-processed using histogram normalization. AlexNet, VGG16 and GoogLeNet CNN classifiers are trained on these images; and the GoogLeNet has achieved 95% recall for binary classification and 75% recall for 3-class classification.
In [15,16,17,18] 5-class classification of DR is performed on Kaggle dataset, however, they use conventional CNNs and/or transfer learning based approaches. In [15] 30000 coloured retinal fundus images are used for training. The images are augmented, normalized and denoised as a pre-processing step. Generated features are fed to award winning CNN architecture for 5-class classification and claimed accuracy upto 85% when validated on 3000 images. In [16], colour normalized and augmented 80000 retinal fundus images are trained on the model and the model has been validated on 5000 images with accuracy of 75%. Retinal fundus images are pre-processed using non-local means denoising (NLMD) for denoising and augmented for the class balance in [17]. For 5-class classification 22.7%, 40%, 41% are achieved by the Baseline model, AlexNet and GoogLeNet respectively. A pre-trained Inception-v3 network is tested for 5-class classification of DR which achieved an accuracy of 48.2% using transfer learning in [18].
3 Data Description and Proposed Pre-processing
In this paper, we have used publicly available Kaggle dataset provided by EyePACS organization [13]. This dataset consists of 35126 retinal fundus images of the left and right eyes. In this experiment, we have randomly chosen 20000 images for training and 3000 images for the test. Average size of each image is \(3000\times 2000\) pixels. The statistics of the DR levels among the images available in Kaggle dataset is listed in Table 1.
3.1 Proposed Pre-processing
This is a crucial step in classification as it greatly helps in the final decision. We propose a three-step pre-processing on the given dataset as follows:
Data Augmentation: Referring to Table 1, one can see that the given dataset has class imbalance, i.e., 73.50% of images are having the class ’No DR’ while remaining class images are 26.50%. Though this is the natural statistics of the data, however, it often introduces bias while training a model. To avoid this, we augment the data (label 1, 2, 3, and 4) by flipping (vertically and horizontally) and rotating images at 45\(^{\circ }\), 90\(^{\circ }\), 135\(^{\circ }\), and 180\(^{\circ }\) in order to match with data label 0. This turns the total 30000 training images with equal class proportion as shown in Table 2.
Corrections Against Sensor Parameters: Typically the retinal fundus images are sensitive to the camera settings including sensor resolution, and also affected by different degrees of noise due to the lightening effects. To mitigate the lightening effects, local average colour of an image is subtracted so that it gets mapped to 50% gray level. This is achieved by first passing the image through a Gaussian filter and then the image is blended. We perform this step as,
where I is the filtered image, \(I_1\) is the given image that should be overlyed on image \(I_2\) which is a Gaussian blurred image, \(\alpha \) and \(\beta \) are transparency levels, and \(\gamma \) is a scalar. In order to create this intermediate image, the images are first scaled to have same radius (300 pixels), then Eq. (1) is modified as \(I = 4I_1 - 4I_2 + 128\). By choosing \(\alpha = 4\) and \(\beta = -4\) we ensure removal of background while extracting foreground features. A scalar \(\gamma = 128\) is added to the weighted sum of two images in order to map local average colour to 50% gray.
Removal of Boundaries:
It is a well-known fact that generally boundaries do not help in extracting features from images. Thus we remove the boundaries of the retinal images by clipping it to 95%. Resultant size of images is \(224 \times 224\) pixels.
Now we present results of the proposed pre-processing steps in some challenging cases in Fig. 1. The left panel of Fig. 1(a) shows a dark image with a poor visual quality which is converted to a better contrast image after the pre-processing as shown in the right panel. In Fig. 1(b) an image (left panel) is shown which is overexposed to light and after the pre-processing (right panel) it is converted to 50% gray level. In Fig. 1(c) noisy image (left panel) with different colour effects is denoised (right panel). Finally Fig. 1(d) shows an image in which foreground features in the given image (left panel) are highlighted in the processed image (right panel) after reducing background effects.
4 Proposed All-CNN for 5-Class DR Detection
In this section, we present our proposed deep All-CNN architecture for the 5-class classification of diabetic retinopathy using retinal fundus images. The All-CNN architecture is inspired from [12].
4.1 Proposed All-CNN
Figure 2 shows the proposed All-CNN architecture. It has 10 convolutional layers and a softmax layer for the 5-class classification. Note that in the proposed model, pooling and fully connected layers are modified by convolutional layers. Increased convolutional layers are perceived to enable the network to learn deeper structures. Hence, it unifies the entire model as convolution operation leading to a simpler architecture. Rectified linear units (ReLu) activation is used at each convolutional layer for incorporating non-linearity in the model. See that the coloured (RGB) pre-processed images (discussed in Sect. 3) of size \(224\times 224\) are given as the input.
In the first convolution layer 32 filters (or kernels) each of size \(7 \times 7\) are used. Since features in retina are wide spreaded, we use large kernel size in order to extract the local dependencies. Following this, second convolution layer with 32 filters each of size \(5 \times 5\) are used. Convolution layer 3 is acting as a pooling layer. We use kernel size \(3 \times 3\) at strides of 2 (performs overlapping convolutions) for dimensionality reduction. Mostly features like exudates and microaneurysms are present nearby. So overlapping convolutions capture such features. Further, fourth convolution layer with 64 number of \(3 \times 3\) filters is used. This is followed by the fifth convolution layer with 64 number of \(3 \times 3\) filters at strides of 2, which acts as another pooling layer. Sixth convolution layer has 128 number of \(3 \times 3\) filters. This is followed by seventh convolution layer with 128 number of \(3 \times 3\) filters at strides of 2, which again acts as a pooling layer. In eighth convolution layer we use 128 number of \(3 \times 3\) filters. Finally, ninth and tenth convolution layers are incorporated to work as the fully connected layers. Ninth convolution layer with 128 number of \(1 \times 1\) filters at strides of 1 and tenth convolution layer with 5 number of \(1 \times 1\) filters at strides of 1 are used. Finally, output from these layers is given to the softmax layer that provides five class predictions corresponding to the DR scale (Fig. 2). Batch normalization is added after each convolutional layer to increase the stability of network. Hence each layer now independently learn features from other layers and thus learn deeper features. Dropout with probability 0.5 is added after the batch normalization at each convolutional layer so that overfitting is avoided and the network is regularized.
Figure 3 shows saliency maps generated at each convolutional layer of the proposed All-CNN (Fig. 2). As shown in Fig. 3, saliency maps allow the medical diagnosis through a detailed visual inspection. They are generated by manipulating gradients of the data that are formed in forward and backward passes. Gradient of the output category is computed with respect to the input image. In Fig. 3 one can see that the gradients are highlighted showing the presence of intrinsic features like microaneurysms, soft exudates and hard exudates which are responsible for the DR levels.
4.2 Training/Test
A supervised training is performed using the proposed architectures shown in Fig. 2 for the 5-class classification of DR. The model uses the cross entropy as loss in the objective function. Stochastic gradient descent optimization is used to minimize the discrepancy between the desired and estimated output of the network. The weights are updated using the backpropogation algorithm. Once the training is done, test images are applied as input for the multi-class classification.
5 Results
In this section, we present the results obtained by proposed architecture on Kaggle dataset. Experiments are conducted on Nvidia Tesla k80 machine with standard memory of 14GB RAM. We make use of 30000 images for training and 3000 images for the test from Kaggle dataset. We run our proposed All-CNN architecture for 200 epochs with a batch size 32 and achieved the accuracy of 86.64% and loss of 0.46. We first calculate confusion matrix and F1 score on the test data. We calculate Recall, Precision, F1 score (Table 3: left panel) using the values in confusion matrix (Table 3: right panel). One can observe that we achieve average F1 score of 0.6318 over the 5-class classification on the test data.
Classification results of the proposed methods and the comparative performance with state-of-art are listed in Table 4. Our proposed architecture performs better detection when compared to other algorithms. It should be noted that algorithms in [15, 16] for 5-class classification use large number of convolutional, Maxpooling and fully connected layers when compared to our proposed All-CNN architecture. Improved pre-processing by the proposed 3-steps and better classification by the proposed All-CNN classifier enable better results.
6 Conclusion
We present a novel deep All-CNN architecture for detection of diabetic retinopathy at the five severity scales. The proposed pre-processing extracts intrinsic features leading to insights for different stages in the disease. Our approach outperformed existing approaches in terms of accuracy, loss, and F1 score. Hence, we have developed a simpler and effective architecture that is useful for practical applications in medical imaging.
References
Tajunisah, I., et al.: Awareness of eye complications and prevalence of retinopathy in the first visit to eye clinic among type 2 diabetic patients. Int. J. Ophthalmol. 4(5), 519–524 (2011)
Cockram, C.S.: The epidemiology of diabetes mellitus in the Asia-Pacific region. Hong Kong Med. J. 6(1), 43–52 (2000)
Wild, S., et al.: Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diab. Care 27(5), 1047–1053 (2004)
The Second National Health and Morbidity Survey: Diabetes mellitus among adults aged 30 years and above, p. 2008. Institute of Public Health (IPH), Ministry of Health, Malaysia (2007)
Kertes, P.J., Johnson, T.M. (eds.): Evidence Based Eye Care. John Wiley and Sons, Lippincott Williams & Wilkins (2007)
Gulshan, V, et al.: Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. Jama, 316(22), 2402–2410 (2016)
Sohini, R., et al.: DREAM: diabetic retinopathy analysis using machine learning. IEEE J. Biomed. Health Inf. 18(5), 1717–1728 (2013)
Lin, Z., et al.: A framework for identifying diabetic retinopathy based on anti-noise detection and attention-based fusion. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11071, pp. 74–82. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00934-2_9
Vogl, W.-D., et al.: Spatio-temporal signatures to predict retinal disease recurrence. In: Ourselin, S., Alexander, D.C., Westin, C.-F., Cardoso, M.J. (eds.) IPMI 2015. LNCS, vol. 9123, pp. 152–163. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-19992-4_12
Cao, C., et al.: Deep learning and its applications in biomedicine. Genomics, Proteomics Bioinf. 16(1), 17–32 (2018)
Litjens, G., et al.: A survey on deep learning in medical image analysis. Med. Image Anal. 42, 60–88 (2017)
Springenberg, J., et al.: Striving for simplicity: the all convolutional net. arXiv preprint arXiv:1412.6806 (2014)
https://www.kaggle.com/c/diabetic-retinopathy-detection/data. Accessed Jan 2017
Lam, C., et al.: Automated detection of diabetic retinopathy using deep learning. In: 2018 AMIA Summits on Translational Science Proceedings, pp. 147–155 (2017)
Ghosh, R., et al.: Automatic detection and classification of diabetic retinopathy stages using CNN. In: 2017 4th International Conference on Signal Processing and Integrated Networks (SPIN), pp. 550–554 (2017)
Pratt, H., et al.: Convolutional neural networks for diabetic retinopathy. In: International Conference On Medical Imaging Understanding and Analysis (MIUA) 2016, vol. 90, pp. 200–205, December 2016
Marco Alban Stanford. Automated detection of diabetic retinopathy using fluorescein angiography photographs, technical report, stanford university (2016)
Masood, S., et al.: Identification of diabetic retinopathy in eye images using transfer learning. In: 2017 International Conference on Computing, Communication and Automation (ICCCA), pp. 1183–1187 (2017)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Challa, U.K., Yellamraju, P., Bhatt, J.S. (2019). A Multi-class Deep All-CNN for Detection of Diabetic Retinopathy Using Retinal Fundus Images. In: Deka, B., Maji, P., Mitra, S., Bhattacharyya, D., Bora, P., Pal, S. (eds) Pattern Recognition and Machine Intelligence. PReMI 2019. Lecture Notes in Computer Science(), vol 11941. Springer, Cham. https://doi.org/10.1007/978-3-030-34869-4_21
Download citation
DOI: https://doi.org/10.1007/978-3-030-34869-4_21
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-34868-7
Online ISBN: 978-3-030-34869-4
eBook Packages: Computer ScienceComputer Science (R0)