Abstract
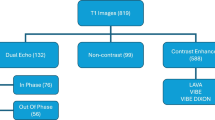
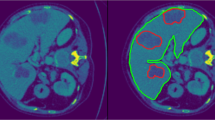
Segmentation of medical images is typically one of the first and most critical steps in medical image analysis. Manual segmentation of volumetric images is labour-intensive and prone to error. Automated segmentation of images mitigates such issues. Here, we compare the more conventional registration-based multi-atlas segmentation technique with recent deep-learning approaches. Previously, 2D U-Nets have commonly been thought of as more appealing than their 3D versions; however, recent advances in GPU processing power, memory, and availability have enabled deeper 3D networks with larger input sizes. We evaluate methods by comparing automated liver segmentations with gold standard manual annotations, in volumetric MRI images. Specifically, 20 expert-labelled ground truth liver labels were compared with their automated counterparts. The data used is from a liver cancer study, HepaT1ca, and as such, presents an opportunity to work with a varied and challenging dataset, consisting of subjects with large anatomical variations responding from different tumours and resections. Deep-learning methods (3D and 2D U-Nets) proved to be significantly more effective at obtaining an accurate delineation of the liver than the multi-atlas implementation. 3D U-Net was the most successful of the methods, achieving a median Dice score of 0.970. 2D U-Net and multi-atlas based segmentation achieved median Dice scores of 0.957 and 0.931, respectively. Multi-atlas segmentation tended to overestimate total liver volume when compared with the ground truth, while U-Net approaches tended to slightly underestimate the liver volume. Both U-Net approaches were also much quicker, taking around one minute, compared with close to one hour for the multi-atlas approach.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Notes
- 1.
(t = 4.886, p = 0.0001) excluding the MAS outlier.
- 2.
(t = 3.499, p = 0.003) excluding the MAS outlier.
- 3.
(t = 4.906, p = 0.0001) excluding the MAS outlier.
- 4.
(t = 5.381, p = 0.00004) excluding the MAS outlier.
References
Lecun, Y., Bengio, Y., Hinton, G.: Deep learning. Nature 521, 436–444 (2015)
Heimann, T., Meinzer, H.P.: Statistical shape models for 3D medical image segmentation: a review. Med. Image Anal. 13, 543–563 (2009)
Fritscher, K., Magna, S., Magna, S.: Machine-learning based image segmentation using Manifold Learning and Random Patch Forests. In: Imaging and Computer Assistance in Radiation Therapy (ICART) Workshop, MICCAI 2015, pp. 1–8 (2015)
Rohlfing, T., Russakoff, D.B., Maurer Jr., C.R.: An expectation maximization-like algorithm for multi-atlas multi-label segmentation. In: Proceedings of the Bildverarbeitung frdie Medizin, pp. 348–352 (2004)
Iglesias, J.E., Sabuncu, M.R.: Multi-atlas segmentation of biomedical images: a survey. Med. Image Anal. 24, 205–219 (2015)
Jorge Cardoso, M., et al.: STEPS: similarity and truth estimation for propagated segmentations and its application to hippocampal segmentation and brain parcelation. Med. Image Anal. 17, 671–684 (2013)
Lecun, Y., Jackel, L.D., Boser, B., Denker, J.S., Gral, H., Guyon, I.: Handwritten digit recognition. IEEE Commun. Mag. 27 (1989)
Zhao, Z.-Q., Zheng, P., Xu, S., Wu, X.: Object detection with deep learning: a review. IEEE Trans. Neural Netw. Learn. Syst. (2019)
Krizhevsky, A., Sutskever, I., Hinton, G.: ImageNet classification with deep convolutional neural networks. In: Advances in Neural Information Processing Systems, vol. 2, pp. 1097–1105 (2012)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 3431–3440 (2015)
Li, X., Chen, H., Qi, X., Dou, Q., Fu, C., Heng, P.: H-DenseUNet: hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Trans. Med. Imaging 37, 2663–2674 (2018)
Gotra, A., et al.: Liver segmentation: indications, techniques and future directions. Insights Imaging 8, 377–392 (2017)
Mole, D.J., et al.: Study protocol: HepaT1ca, an observational clinical cohort study to quantify liver health in surgical candidates for liver malignancies. BMC Cancer 18, 890 (2018)
Avants, B.B., Tustison, N.J., Song, G., Cook, P.A., Klein, A., Gee, J.C.: A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 54(3), 2033–2044 (2010)
Heinrich, M.P., Jenkinson, M., Brady, S.M., Schnabel, J.A.: Globally optimal deformable registration on a minimum spanning tree using dense displacement sampling. In: Ayache, N., Delingette, H., Golland, P., Mori, K. (eds.) MICCAI 2012. LNCS, vol. 7512, pp. 115–122. Springer, Heidelberg (2012). https://doi.org/10.1007/978-3-642-33454-2_15
Xu, Z., et al.: Evaluation of six registration methods for the human abdomen on clinically acquired CT. IEEE Trans. Biomed. Eng. 63, 1563–1572 (2016)
Irving, B., et al.: Deep quantitative liver segmentation and vessel exclusion to assist in liver assessment. In: Valdés Hernández, M., González-Castro, V. (eds.) MIUA 2017. CCIS, vol. 723, pp. 663–673. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-60964-5_58
Ioffe, S., Szegedy, C.: Batch normalization: accelerating deep network training by reducing internal covariate shift (2015)
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., Wojna, Z.: Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2818–2826 (2016)
Tustison, N.J., et al.: N4ITK: improved N3 bias correction. IEEE Trans. Med. Imaging 29, 1310–1320 (2010)
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. arXiv Preprint arXiv:1412.6980 (2014)
Antonelli, M., et al.: GAS: a genetic atlas selection strategy in multi-atlas segmentation framework. Med. Image Anal. 52, 97–108 (2019)
Acknowledgements
Portions of this work were funded by a Technology Strategy Board/Innovate UK grant (ref 102844). The funding body had no direct role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Owler, J., Irving, B., Ridgeway, G., Wojciechowska, M., McGonigle, J., Brady, S.M. (2020). Comparison of Multi-atlas Segmentation and U-Net Approaches for Automated 3D Liver Delineation in MRI. In: Zheng, Y., Williams, B., Chen, K. (eds) Medical Image Understanding and Analysis. MIUA 2019. Communications in Computer and Information Science, vol 1065. Springer, Cham. https://doi.org/10.1007/978-3-030-39343-4_41
Download citation
DOI: https://doi.org/10.1007/978-3-030-39343-4_41
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-39342-7
Online ISBN: 978-3-030-39343-4
eBook Packages: Computer ScienceComputer Science (R0)