Abstract
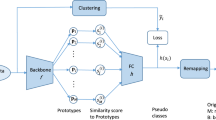
The recent success of convolutional neural networks (CNNs) attracts much attention to applying a computer-aided diagnosis system for digital pathology. However, the basis of CNN’s decision is incomprehensible for humans due to its complexity, and this will reduce the reliability of its decision. We improve the interpretability of the decision made using the CNN by presenting them as co-occurrences of interpretable components which typically appeared in parts of images. To this end, we propose a prototype-based interpretation method and define prototypes as the components. The method comprises the following three approaches: (1) presenting typical parts of images as multiple components, (2) allowing humans to interpret the components visually, and (3) making decisions based on the co-occurrence relation of the multiple components. Concretely, we first encode image patches using the encoder of a variational auto-encoder (VAE) and construct clusters for the encoded image patches to obtain prototypes. We then decode prototypes into images using the VAE’s decoder to make the prototypes visually interpretable. Finally, we calculate the weighted combinations of the prototype occurrences for image-level classification. The weights enable us to ascertain the prototypes that contributed to decision-making. We verified both the interpretability and classification performance of our method through experiments using two types of datasets. The proposed method showed a significant advantage for interpretation by displaying the association between class discriminative components in an image and the prototypes.
The part of this work is based on results obtained from a project commissioned by the New Energy and Industrial Technology Development Organization (NEDO).
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Adadi, A., Berrada, M.: Peeking inside the black-box: a survey on explainable artificial intelligence (XAI). IEEE Access 6, 52138–52160 (2018)
Arandjelović, R., Gronat, P., Torii, A., Pajdla, T., Sivic, J.: NetVLAD: CNN architecture for weakly supervised place recognition. In: IEEE Conference on Computer Vision and Pattern Recognition (2016)
Chen, C., Li, O., Barnett, A., Su, J., Rudin, C.: This looks like that: deep learning for interpretable image recognition. arXiv arXiv:1806.10574 (2018)
Csurka, G., Dance, C.R., Fan, L., Willamowski, J., Bray, C.: Visual categorization with bags of keypoints. In: Workshop on Statistical Learning in Computer Vision, ECCV, pp. 1–22 (2004)
Bejnordi, B.E., et al.: Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 318(22), 2199–2210 (2017). https://doi.org/10.1001/jama.2017.14585. The CAMELYON16 Consortium machine learning detection of breast cancer lymph node metastases
Erhan, D., Bengio, Y., Courville, A., Vincent, P.: Visualizing higher-layer features of a deep network. Tech. rep. 1341, University of Montreal (June 2009)
Ioffe, S., Szegedy, C.: Batch normalization: accelerating deep network training by reducing internal covariate shift. In: Proceedings of the 32nd International Conference on Machine Learning, vol. PMLR37, pp. 448–456 (2015)
Kingma, D.P., Welling, M.: Auto-encoding variational bayes. In: 2nd International Conference on Learning Representations, ICLR 2014, Banff, AB, Canada, April 14–16, 2014, Conference Track Proceedings (2014). http://arxiv.org/abs/1312.6114
Kylberg, G.: The Kylberg texture dataset v. 1.0. External report (blue series) 35, Centre for Image Analysis, Swedish University of Agricultural Sciences and Uppsala University, Uppsala, Sweden (September 2011). http://www.cb.uu.se/~gustaf/texture/
LeCun, Y., Bengio, Y., Hinton, G.: Deep learning. Nature 521, 436–444 (2015)
Li, O., Chen, H.L.C., Rudin, C.: Deep learning for case-based reasoning through prototypes: a neural network that explains its predictions. In: Proceedings of AAAI, pp. 123–456 (2018)
Liu, Y., et al.: Detecting cancer metastases on gigapixel pathology images. Tech. rep. arXiv arXiv:1703.02442 (2017)
Selvaraju, R.R., Cogswell, M., Das, A., Vedantam, R., Parikh, D., Batra, D.: Grad-cam: visual explanations from deep networks via gradient-based localization. In: International Conference on Computer Vision (ICCV). IEEE (2017)
Simonyan, K., Vedaldi, A., Zisserman, A.: Deep inside convolutional networks: visualising image classification models and saliency maps. CoRR arXiv:1312.6034 (2013)
Veeling, B.S., Linmans, J., Winkens, J., Cohen, T., Welling, M.: Rotation equivariant CNNs for digital pathology. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11071, pp. 210–218. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00934-2_24
Zeiler, M.D., Fergus, R.: Visualizing and understanding convolutional networks. In: Fleet, D., Pajdla, T., Schiele, B., Tuytelaars, T. (eds.) ECCV 2014. LNCS, vol. 8689, pp. 818–833. Springer, Cham (2014). https://doi.org/10.1007/978-3-319-10590-1_53
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Uehara, K., Murakawa, M., Nosato, H., Sakanashi, H. (2020). Prototype-Based Interpretation of Pathological Image Analysis by Convolutional Neural Networks. In: Palaiahnakote, S., Sanniti di Baja, G., Wang, L., Yan, W. (eds) Pattern Recognition. ACPR 2019. Lecture Notes in Computer Science(), vol 12047. Springer, Cham. https://doi.org/10.1007/978-3-030-41299-9_50
Download citation
DOI: https://doi.org/10.1007/978-3-030-41299-9_50
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-41298-2
Online ISBN: 978-3-030-41299-9
eBook Packages: Computer ScienceComputer Science (R0)