Abstract
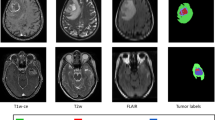
We propose a deep learning based approach for automatic brain tumor segmentation utilizing a three-dimensional U-Net extended by residual connections. In this work, we did not incorporate architectural modifications to the existing 3D U-Net, but rather evaluated different training strategies for potential improvement of performance. Our model was trained on the dataset of the International Brain Tumor Segmentation (BraTS) challenge 2019 that comprise multi-parametric magnetic resonance imaging (mpMRI) scans from 335 patients diagnosed with a glial tumor. Furthermore, our model was evaluated on the BraTS 2019 independent validation data that consisted of another 125 brain tumor mpMRI scans. The results that our 3D Residual U-Net obtained on the BraTS 2019 test data are Mean Dice scores of 0.697, 0.828, 0.772 and Hausdorff\(_{95}\) distances of 25.56, 14.64, 26.69 for enhancing tumor, whole tumor, and tumor core, respectively.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Kamnitsas, K., et al.: Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. MIA 36, 61–78 (2017)
Isensee, F., Kickingereder, P., Wick, W., Bendszus, M., Maier-Hein, K.H.: Brain tumor segmentation and radiomics survival prediction: contribution to the BRATS 2017 challenge. In: Crimi, A., Bakas, S., Kuijf, H., Menze, B., Reyes, M. (eds.) BrainLes 2017. LNCS, vol. 10670, pp. 287–297. Springer, Cham (2018). https://doi.org/10.1007/978-3-319-75238-9_25
Li, X., Chen, H., Qi, X., Dou, Q., Fu, C.-W., Heng, P.A.: H-DenseUNet: hybrid densely connected UNet for liver and liver tumor segmentation from CT volumes. arXiv preprint arXiv:1709.07330 (2017)
Isensee, F., Jaeger, P.F., Full, P.M., Wolf, I., Engelhardt, S., Maier-Hein, K.H.: Automatic cardiac disease assessment on cine-MRI via time-series segmentation and domain specific features. In: Pop, M., et al. (eds.) STACOM 2017. LNCS, vol. 10663, pp. 120–129. Springer, Cham (2018). https://doi.org/10.1007/978-3-319-75541-0_13
Kamnitsas, K., et al.: Ensembles of multiple models and architectures for robust brain tumour segmentation. In: Crimi, A., Bakas, S., Kuijf, H., Menze, B., Reyes, M. (eds.) BrainLes 2017. LNCS, vol. 10670, pp. 450–462. Springer, Cham (2018). https://doi.org/10.1007/978-3-319-75238-9_38
Bakas, S., et al.: Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Nat. Sci. Data 4, 170117 (2017)
Bakas, S., et al.: Segmentation labels and radiomic features for the pre-operative scans of the TCGA-GBM collection. The Cancer Imaging Archive 286 (2017)
Bakas, S., et al.: Segmentation labels and radiomic features for the pre-operative scans of the TCGA-LGG collection. The Cancer Imaging Archive (2017)
Menze, B.H., Jakab, A., Bauer, S., et al.: The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging 34(10), 1993–2024 (2015). https://doi.org/10.1109/TMI.2014.2377694
Bakas, S., Reyes, M., Jakab, A., Bauer, S., Rempfler, M., Crimi, A., et al.: Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. arXiv preprint arXiv:1811.02629 (2018)
Çiçek, Ö., Abdulkadir, A., Lienkamp, S.S., Brox, T., Ronneberger, O.: 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 424–432. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_49
Arora, R., Basu, A., Mianjy, P., Mukherjee, A.: Understanding deep neural networks with rectified linear units. arXiv preprint arXiv:1611.01491 (2016)
Ulyanov, D., Vedaldi, A., Lempitsky, V.: Instance normalization: the missing ingredient for fast stylization. arXiv preprint arXiv:1607.08022 (2016)
Salehi, S.S.M., Erdogmus, D., Gholipour, A.: Tversky loss function for image segmentation using 3D fully convolutional deep networks. In: Wang, Q., Shi, Y., Suk, H.-I., Suzuki, K. (eds.) MLMI 2017. LNCS, vol. 10541, pp. 379–387. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-67389-9_44
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Bhalerao, M., Thakur, S. (2020). Brain Tumor Segmentation Based on 3D Residual U-Net. In: Crimi, A., Bakas, S. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2019. Lecture Notes in Computer Science(), vol 11993. Springer, Cham. https://doi.org/10.1007/978-3-030-46643-5_21
Download citation
DOI: https://doi.org/10.1007/978-3-030-46643-5_21
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-46642-8
Online ISBN: 978-3-030-46643-5
eBook Packages: Computer ScienceComputer Science (R0)