Abstract
The mining of time series data plays an important role in modern information retrieval and analysis systems. In particular, the identification of similarities within and across time series has garnered significant attention and effort over the last few years. For this task, the class of matrix profile algorithms, which create a generic structure that encodes correlations among records and dimensions—the matrix profile—is a promising approach, as it allows simplified post-processing and analysis steps by examining the resulting matrix profile structure. However, it is expensive to create a matrix profile: it requires significant computational power to evaluate the distance among all subsequence pairs in a time series, especially for very long and multi-dimensional time series with a large dimensionality. Existing approaches are limited in their scalability, as they do not target High Performance Computing systems, and—for most realistic problems—are suited only for datasets with a small dimensionality.
In this paper, we introduce a novel MPI-based approach for the calculation of a matrix profile for multi-dimensional time series that pushes these limits. We evaluate the efficiency of our approach using an analytical performance model combined with experimental data. Finally, we demonstrate our solution on a 128-dimensional time series dataset of 1 million records, solving 274 trillion sorts at a sustained 1.3 Petaflop/s performance on the SuperMUC-NG system.
D. Yang—This research is completed at TU Munich.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
1 Introduction
State-of-the-art physical systems, such as monitoring infrastructures or operational logs of industrial machines, often generate a time-tagged series of data points in a given order. A collection of such data points over time, which is called a time series, is crucial to understanding the underlying behavior of the physical system that produces it. Such time series are usually provided in the form of a collection of individual time series, which together form a multi-dimensional time series.
One important aspect in understanding multi-dimensional time series is the explorative discovery of similar and repeating patterns in a (potentially large) dataset [4]. Recent advances in data mining techniques enable the extraction of complex pattern structures in multi-dimensional time series. They generally rely on computing generic similarity data structures, e.g., correlation information from the individual time series.
One prominent example for time series data mining is the matrix profile approach, which has been introduced by Yel et al. [24] and has been successfully applied to many datasets from various fields. A matrix profile is a generic meta series, i.e., a time series itself that provides information about the input series by summarizing correlations and nearest neighbor indices among subsequences in a set of given time series. A matrix profile also enables easier in-depth studies of patterns and anomalies, and with that many data mining tasks, such as the discovery of frequent patterns, correlations, and clusters in a dataset.
State-of-the-art algorithms for the computation of a matrix profile mostly target one-dimensional time series, i.e., a single time series covering one sensor input. Only recently, the first algorithms for multi-dimensional time series appeared [23] offering detailed insights into repeating patterns across different time series, significantly increasing the ability to understand multi-dimensional time series. However, these approaches are significantly more compute-intensive than one-dimensional matrix profile algorithms and hence are no longer feasible on standard systems, which they currently target, for realistic workloads. They have not been shown to scale to larger systems nor that they can be used on anything but small datasets.
However, multi-dimensional time series with large numbers of records in each time series are typical in many disciplines. One example is the operation of industrial gas turbines. Such systems are monitored by more than 100 different sensors and generate millions of recordsFootnote 1 per month [10]. The analysis of this data can generate new insights about correlations among different sensors and operational modes, which can be used to optimize the operation of a gas turbine for more stable operation, better fuel efficiency, and consequently less air pollution. Another real-world example is monitoring of HPC infrastructures. Netti et al. [14] use up to 3176 sensors per node to monitor multiple production HPC systems at the Leibniz Supercomputing Centre and have shown that the collected monitoring data contains valuable information on the system’s behavior, and can be used, eg., in the characterization of applications running on it.
To apply the concept of the matrix profile to large-scale multi-dimensional time series and hence to such real-world problems, we require new approaches to scale the computation of matrix profiles both to larger computational resources and to larger datasets. In this work, we provide a novel approach of calculating the matrix profile for large multi-dimensional time series in parallel on HPC systems. We build our approach on the observation that the calculation of a matrix profile is highly memory bound [15], and therefore can benefit from horizontal scaling of memory bandwidth and throughput. In addition, parallel computation of the matrix profile requires the aggregation of final results, i.e., a series of reduction operations, which can exploit high performance interconnects. However, in order to achieve efficiency, a series of algorithmic advances and optimization steps are needed, which we introduce in this work and verify with an analytical performance model.
In particular, the contributions in this paper are:
-
We introduce a new highly-parallel algorithm to compute the matrix profile for multi-dimensional time series.
-
We provide an analytical and experimental model for the performance of our algorithm.
-
We provide a scalable implementation of our algorithm to compute a multi-dimensional matrix profile efficiently on a Petascale HPC system.
Using our novel algorithmic approach, we demonstrate the computation on a 128-dimensional time series dataset of 1 million records on the SuperMUC-NG Petascale system, solving 274 trillion sorts at sustained 1.3 Petaflop/s performance.
The remainder of this paper is organized as follows: Sect. 2 introduces the concept of matrix profile, existing algorithms to compute it, and the challenges to running this computation on HPC systems. Section 3 discusses our approach for parallel computation of the matrix profile. Section 4 describes our MPI implementation, and Sect. 5 presents a performance model to describe the workload of matrix profile computation for a multi-dimensional time series on a parallel system. Section 6 explains and analyzes our experiments, Sect. 7 discusses related work, and Sect. 8 provides conclusions and final discussions.
2 Background on Matrix Profile
A matrix profile is a generic similarity indexing approach for the analysis of one- or multi-dimensional time series, in particular, the investigation and quantification of similar patterns. This analysis relies on the study of similarities (or correlations) among local chunks—i.e., subsets of continuous values—of the time series, referred to as subsequences. A matrix profile summarizes a complete correlation matrix (or equivalently distance matrix) of all the subsequences of a time series, the distance matrix, into mainly two data structures:
-
A matrix profile P, which is a meta series encoding the distance of a subsequence to its nearest neighbor, and
-
A matrix profile index I, which is an indexing structure storing pointers to the nearest neighbor of a subsequence in the time series.
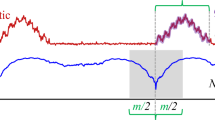
Illustration of automatic semantic segmentation using a multi-dimensional matrix profile. We show a synthetic time series of three sensors, the corresponding matrix profile, and we group the patterns in the time series based on matrix profile into four clusters denoted by colors. Each sensor consists of \(20\,000\) samples. The subsequence length for the analysis is set to 4 s, and we use k-means with 4 clusters on the 3D matrix profile to distinguish the clusters. (Color figure online)
The terminology introduced above, i.e., matrix profile and matrix profile index, in principle, also applies to multi-dimensional time series. However, for a multi-dimensional time series, the notion of similarity is generalized so that the analysis takes the correlation among subsequences in different dimensions into account. Moreover, the resulting matrix profile (and index) for a time series with the dimensionality of d consists of d matrix profiles. These profiles are presented in a single matrix with d rows where the k-th (\(k \le d\)) row of the matrix profile provides information about the nearest neighbors of subsequences in the best matching k dimensions.
To illustrate the matrix profile, we present a simple scenario in Fig. 1 and show how a matrix profile can be used for automatic semantic segmentation of a multi-dimensional time seriesFootnote 2. Our synthetic example uses a set of 3 sensors: the first carries sin(x)-wavelets starting at +20 s; the second carries square-wavelets starting at time +40 s; and the third carries a sawtooth-wavelet starting at +70 s; all wavelets are repeated every 10 s. This introduces four phases in the presented sensor data during which the correlations among the sensors are unique. The generated matrix profile highlights and distinguishes these phases by summarizing the correlation structure among the sensors in the time series.
We perform a multi-dimensional matrix profile analysis on this example. The resulting matrix profile (only the last row in the matrix profile corresponding to the nearest neighbors of 3D patterns) is presented in the lower graph colored in black in Fig. 1. We use a k-means algorithm to cluster this matrix profile and group the corresponding motifs—which are patterns with a unique correlation structure among all three sensors—based on their distances in the matrix profile (see Fig. 1, color-coded time series segments). This analysis results in meaningful clusters corresponding to the respective phases in the original data.
Algorithms for Calculation of Matrix Profile
We base our work on the state-of-the-art algorithm STOMP (Scalable Time Series Ordered-search Matrix Profile) [26], which computes a one-dimensional matrix profile with optimal complexity. This algorithm is later optimized (referred to hereafter as \(STOMP_{opt}\)) for a better arithmetic intensity and numerical stability [27] to enable the calculation of a matrix profile on large (single-dimensional) time series. In particular, STOMP-based algorithms allow a partial storage of the distance matrix—a matrix of, e.g., Euclidean distances among all subsequences—using an ordered iterative solver for matrix profiles by computing a so-called distance profile. The distance profile is one row within the distance matrix (see Eq. 8 and Fig. 2 right), which is computed in a given iteration to update the matrix profile data structure.
For the calculation of a matrix profile over a multi-dimensional time series, Yeh et al. [23] introduce mSTAMP. It uses STOMP-based formulations for the computation of the distance profile. mSTAMP iteratively computes the matrix profile by independent calculation of distance profiles for all dimensions. Unlike STOMP, in each iteration, before updating the resulting intermediate state of the matrix profile, mSTAMP sorts the distance profiles in each record separately. However, this method has three significant drawbacks: (a) it does not exploit the numerically stable formulation for computation of a matrix profile, (b) it is only available as a scripted prototype in Python and Matlab, and (c) it exists only in a sequential version. Consequently, it is currently insufficient for computing a matrix profile on a large-scale multi-dimensional real-world dataset.
3 Multi-dimensional Parallel Matrix Profile: \((MP)^{N}\)
To overcome the limitations stated above, we introduce a new approach to compute multi-dimensional matrix profiles. Building on top of the existing mSTAMP concepts, we introduce \((MP)^{N}\) (stands for Multi-dimensional Parallel Matrix Profile), which is designed to exploit the computational power, high performance interconnect, and I/O capabilities of HPC systems to calculate matrix profiles of multi-dimensional datasets within realistic execution times.
For this, the mSTAMP formulations can be adapted for parallel processing, allowing the distribution of computational workload among multiple workers with minimum communication during the computation phase. By partitioning the time series along records, we can distribute the workload across multiple processing elements, e.g., cores of a multiprocessor. This workload consists of (1) the computation of the distance profile, (2) the sorting of the distance profile, and (3) the updates on the resulting matrix profile. Finally, the results residing in the memory of the various machines are merged by performing reduction operations. This way, we can scale the computation of a multi-dimensional matrix profile to a large number of nodes.
3.1 Overview of the \((MP)^{N}\)Algorithm
The foundation of our algorithm is an adapted version of mSTAMP based on \(STOMP_{opt}\) kernels. Its mathematical representation is given in Table 1, which provides us good numerical stability and a better single thread arithmetic performance for the evaluation of the distance matrix [27].
For a multi-dimensional time series with d dimensions, each of length (number of records) n, and considering a subsequence length m, we evaluate a total of d distance matrices, one for each dimension. A distance matrix is a symmetric matrix storing the Euclidean distance among all subsequence pairs of the input time series separately for every dimension.
In contrast to mSTAMP, we explicitly take the symmetric structure of the distance matrices into account to avoid redundant computations. Further, we consider the fact that there is a significant semantic difference between one- and multi-dimensional motifs: in multi-dimensional motifs, subsequences can correlate with any other subsequences in any k-dimensional (\(k \le d\)) subspace across all dimensions. This results in a matrix profile that is defined as the minimum value of each column in the distance matrices after sorting and partial aggregation of resulting distance profiles of the d dimensions. The resulting matrix profile represents the nearest neighbors of a subsequence with the best matching k (\(k \le d\)) dimensions. To achieve this with minimal overhead, we sort the distances across the dimensions for all records in the distance profile using an optimized memory layout combined with a high-performance sorting kernel. Accordingly, the matrix profile index is defined as the argminFootnote 3 pointing to the closest neighbor of each subsequence (Eq. 10).
In the case of a self-joinFootnote 4, we exclude the trivial matches, i.e., a subsequence matching a given region including itself or its neighboring subsequences. These regions correspond to the proximities of the diagonal entries in the distance matrices. \((MP)^{N}\) invalidates these trivial matches in so-called “exclusion zones” when merging the calculated intermediate profiles into the final result.
At the end of each iteration, \((MP)^{N}\) merges the sorted and aggregated distance profiles into the final matrix profile and its index using element-wise min and argmin operations (see Eq. 10).

3.2 Iterative Computation of the Matrix Profile
We use the formulas in Table 1 for the iterative computation of the distance matrix and its distance profile (Algorithm 1), which can be summarized in the following steps (see Fig. 2 right):
-
1.
First, we (pre-)compute statistics of all subsequences using Eqs. 1–4 in all d dimensions (Line 1, Algorithm 1).
-
2.
In Line 2, Algorithm 1, we initialize the streaming dot product (see the formulation developed by Zimmerman et al. [27]) with the distance between the first subsequence in the time series and all other subsequences and (pre-)compute the distance profiles for all d dimensions.
-
3.
We use Eqs. 5–8 to iteratively—and in-situ—update the distance profile \(\delta \) (Line 3, Algorithm 1), by calculating the Euclidean distances between subsequence idx and all the other subsequences i. This, in turn, is done for all d dimensions (Lines 4–8, Algorithm 1). Further, in each iteration idx, we\(\dots \)
-
(a)
\(\dots \)invalidate the entries in the exclusion zone of \(\delta \) (Line 9, Algorithm 1).
-
(b)
\(\dots \)sort the distance profile \(\delta \) in each record and across the dimensions (Eq. 9 and Lines 10–12, Algorithm 1).
-
(c)
\(\dots \)combine the sorted distance profile with the matrix profile for all d dimensions using element-wise min and argmin operations, i.e., the matrix profile and its index are updated according to the distance profile (Eq. 10, Lines 13–18, Algorithm 1 and Fig. 2 right).
-
(a)
3.3 Algorithmic Optimizations in \((MP)^{N}\)
As discussed in Sect. 3.1, we only compute the upper triangular part of the distance matrix and then compensate for this by updating the matrix profile accordingly. For that, we introduce an additional set of matrix profiles \(P_c\) and matrix profile indices \(I_c\). \(P_c\) and \(I_c\) represent column-wise (for column c) matrix profiles and indices (similar to the definitions by Zimmerman [27]), and are both multi-dimensional (Fig. 2 right). We compute the partial results \(P_c\) and \(I_c\) from already computed distance profiles using the transpose of the distance matrix instead of computing the lower triangular part of the distance matrix. We then compute \(P_c\) and \(I_c\) using min and argmin operations per dimension at each iteration on the distance profile \(\delta \). Finally, we construct the final matrix profile and its index by merging the results of \(P_c\) to P, and \(I_c\) to I (Line 20, Algorithm 1). This optimization and merging scheme are parallelizable, as we demonstrate below in our MPI implementation of \((MP)^{N}\).
3.4 Data Layout Considerations
For optimal data-locality, we use a column-wise in-memory data layout for most data structures, as most kernels access the data structures in a column-wise data layout (see Fig. 2 left). Only for the distance profile \(\delta \), we use a double buffering scheme, where data is accessed row-wise as well. This way, vectorized sorting kernels can exploit a more efficient access pattern to \(\delta \).
Illustration of the column- and row-wise layouts for storage of the distance profile \(\delta \) (left). Each column with a unique color represents one dimension, and rows represent records. Besides, the red arrows indicate the data layout. On the right, the iterative computation of the matrix profile in all dimensions is visualized. The active distance profiles in a specific iteration are colored in green. These two distance profiles are sorted and partially aggregated, and the corresponding elements in the matrix profile and its index are updated—colored in brown and magenta. (Color figure online)
3.5 Partitioning and Aggregation Scheme
Apart from the pre-computation of the first row of each local distance matrix, the iterative computation of the distance matrix and matrix profile, as introduced in Sect. 3.2, is an embarrassingly-parallel workload. Therefore, each process has to compute the first iteration using a naive sliding dot product formulation [24] (Fig. 2 right). Figure 3 left illustrates the partitioning scheme used in \((MP)^{N}\) for a two-dimensional problem by decomposing the problem into four independent subproblems. Here, the iterative evaluation of the two distance matrices—one for each dimension—in a two-dimensional problem is distributed among four processing units. Note that \((MP)^{N}\) computes the matrix profile by evaluating only the upper triangular parts of the partitioned distance matrix. In addition, we introduce an additional final step to aggregate the partial results from different processing units (cf. the merging scheme in Sect. 3.3). Due to the symmetry of the distance matrix, the following property for all resulting matrix profiles in the partitions holds: \(P^{(i,j)} = P^{(j,i)}_{c}\)—where (i, j) is the index of a process in the virtual topology. Given this property, the partial matrix profiles are merged according to the following equation (see Fig. 3 left):
By generalizing Eq. 11, we can formulate this merging step for aggregation of partial matrix profiles in partitions (cf. Fig. 3). The merge operation in Eq. 11 represents element-wise reduction similar to Lines 15 and 17, Algorithm 1.
(Left) Illustration for the partitioning scheme of the distance matrix in \((MP)^{N}\) for a two-dimensional matrix profile (cf. Fig. 2): we use four partitions in this illustration. (Right) Aggregation scheme: the parts of the distance matrix with a similar color are computed on the same partition, e.g., partitions colored in pink are computed in the processing element with id (0, 1). (Color figure online)
4 MPI-Based Parallelization and Optimization
We have implemented the parallel version of our \((MP)^{N}\) algorithm using the Message Passing Interface (MPI), which allows us to exploit the performance advantages in tightly coupled HPC systems. In particular, we replace a filesystem-based final aggregation [27] with high-performance reduction operations, utilize fast communication and scalable I/O functionality for massively-parallel pre- and post-processing, and enable unprecedented scaling, all of which have been bottlenecks in existing cloud-based parallel matrix profile algorithms.
4.1 Phases of \((MP)^{N}\)
Our \((MP)^{N}\) implementation includes six phases (see Fig. 4):
-
1.
Setup Phase: We first build a virtual topology of processes (see Fig. 5). Using this topology, we create the MPI communicators required for reading and writing input and output time series from the file system, as well as the MPI communicators required for the distribution of time series and the aggregation of the matrix profile. Finally, we distribute the data and workload among all MPI processes according to our partitioning algorithm (see Sect. 3.5).
-
2.
File Read Phase: We read the time series from the file, using a “row-wise” layout for storing data, i.e., the data points from different data streams with a given timestamp are stored consecutively in the file. This simplifies the partitioned read operation when using a large number of processes.
-
3.
Data Distribution Phase: Processes that participate in the File Read Phase broadcast the input time series via the MPI communicators created during the setup.
-
4.
Kernel Execution Phase: We execute the necessary kernels to compute a local multi-dimensional matrix profile for a subset (partition) of the input series using the iterative algorithm introduced in Sect. 3.
-
5.
Aggregation Phase: We aggregate and merge the final results (see Eq. 11 and Fig. 5). This is done using reduction operations over MPI communicators created in the Setup Phase. After this phase, the final results are available on all processes that participate in the Write Phase.
-
6.
Write Phase: A subset of MPI processes responsible for parallel-write operations outputs the final matrix profile and its index using MPI I/O (Fig. 5).
Illustration of the virtual topology of MPI processes in communicators. Each triangle with a distinct color represents a separate MPI process working on a distinct part of the distance matrix. The red boxes represent exclusive process sets in communicators. The left and middle figures represent communicators used in Data Distribution and Aggregation phases, and the right figure shows the communicator used in I/O phases. The small boxes in each process represent data buffers associated with a local matrix profile and its index to be reduced in the Aggregation phase. Buffers depicted with the same colors within the same process group are reduced. (Color figure online)
4.2 MPI Communicators in \((MP)^{N}\)
We use three communicators in various phases presented in Fig. 4. These communicators are illustrated in Fig. 5 and are as follows:
-
For reading and writing, we use only MPI processes responsible for the diagonals on the virtual topology (Fig. 5 right). This allows us to reduce the pressure on the parallel file system caused by the large amount of I/O operations (IOP), which would otherwise degrade the I/O performance.
-
We use both column- and row-wise communicators for the Distribution of data and the reduction of the final result in the aggregation phase. We further use a custom argmin MPI reduction operation, introduced in Sect. 3, according to the merging scheme described in Sect. 3.5. This custom reduction is executed on the row-wise and column-wise communicators in the Aggregation Phase.
5 Modeling the Performance Bottlenecks
To understand the scaling behavior of our algorithm, we introduce an analytical performance model for \((MP)^{N}\). We base our model on two inputs: the number of processes and the problem size. Our model includes all phases in the computation of our algorithm as introduced in Sect. 4.1. We decompose the execution in shares of execution time for these phasesFootnote 5:
We consider a time series of size n with d dimensions resulting in \(n \times d\) as the input size, and we assume a total of p MPI processes. Using these notations, the following equations describe the scaling behavior of each phase in \((MP)^{N}\):
However, to simplify our discussions regarding the scaling bottlenecks of \((MP)^{N}\), we only present our model for dominant portions of the runtime, which are \(T_{Setup}\) and \(T_{Kernel}\) (cf. Sect. 6.4). We consider a linearithmic growth of \(T_{Setup}\) with respect to p (see Moody et al. [12]), and the growth of \(T_{Kernel}\) is driven from the parent algorithm mSTAMP, and is modified for \((MP)^{N}\). This results in the final model of:
6 Evaluation
To demonstrate the performance of our implementation, we conduct a wide set of experiments. All our experiments are carried out on the SuperMUC-NGFootnote 6 system at the Leibniz Supercomputing Centre (LRZ). Each SuperMUC-NG node features two 24-core Intel Xeon Platinum 8174 Processors (Skylake) running at 2.69 GHz and 96 GB main memory. Our code is implemented in C++ and uses double-precision floating-point values in the kernels. We use the Intel C++ compiler v19.0 update 4.0 for compilation, and unless otherwise noted, we use Intel MPI.
All our scaling experiments are executed using 48 MPI processes per node and each MPI process is mapped to one physical core. Thus, we use the terms cores and processes interchangeably in the remainder of this paper. Moreover, all input sets are randomly generated sequences, as the performance is agnostic to the input data used.
6.1 Correctness and Numerical Stability
To validate the correctness and stability of \((MP)^{N}\), we compare the matrix profile and its indices calculated with different numbers of processes, as sequential baseline executions are infeasible for our target input datasets. For that, we fix the setup to \(n=524\,288\), \(d=128\) and \(m=512\), and execute \((MP)^{N}\) on 512 MPI processes to create a baseline. We then repeat the analysis using \(1\,024\), \(4\,096\), \(16\,384\), and \(65\,536\) processes. In all experiments, \(L_{1}\) and \(L_{\infty }\)Footnote 7 norms of the resulting matrix profiles are accurate on up to 15 significant digits and the matrix profile index fully matches the baseline. This confirms the numerical stability of our solution with respect to double-precision floating-point numbers.
6.2 Single-Core Performance
We evaluate how the problem size parameters and the difference in sorting kernels affect the execution time on a single-core setup. In the following, each experiment is repeated 5 times, and we show the average result.
For the sorting-kernel used during construction of the complete matrix profile, we use existing standard libraries. In particular, we compare three leading libraries for their performance:
-
AVX-512-bitonic [3], which is a high performance sorting kernel for small- and mid-sized arrays optimized for the Intel Skylake architecture
-
Intel Integrated Performance Primitives (IPP) library [19], as a vendor-specific alternative
-
qsort from C++–stdlib-based [9] as a basic Quicksort implementation
First, we analyze the effect of the dimensionality parameter d. Figure 6 top illustrates our result. Different colors represent the sorting kernels used in a given experiment. We confirm an expected linear growth in execution time for all three kernels, which validates our proposed model presented in Sect. 5. While all three kernels complete in similar time for smaller dimensionality, AVX-512-bitonic provides the best performance with an increasing dimensionality, which is our target. The superiority of AVX-512-bitonic is in accordance with the results of an existing study by Bramas et al. [3]. Therefore, we fix the AVX-512-bitonic kernel for all further experiments.
(Top) Linearithmic growth of execution time vs. increasing dimensionality parameter d, with \(n=2^{14}\) samples and \(m=2^{9}\). (Middle) Quadratic growth of execution time vs. increasing number of records n, with \(d=2^5\), \(m=2^{9}\). (Bottom) Near-constant execution time with increasing subsequence length m, with \(n=2^{14}\), \(d=2^{5}\).
Second, we analyze the effect of the number of records n on execution time. Figure 6 middle shows a quadratic growth of time with respect to the number of records matching our model in Sect. 5 as well as a previous study by Yeh et al. [23]. The subsequence length m has a limited effect on execution time (see Fig. 6 bottom), which is also confirmed by Yeh et al. [23].
Overall, we confirm that the single-node performance of our adapted algorithm is preserved in relation to the STOMP-based mSTAMP [23], and we validate our execution time model against the obtained experimental data.
6.3 Single-Node Performance
Next, we present results from single-node execution. We execute \((MP)^{N}\) with I_MPI_PIN_ORDER=scatter to pin the MPI processes within NUMA nodesFootnote 8. This ensures optimal sharing of the available memory bandwidth across all cores.
As shown in Fig. 7, we can see the expected performance saturation pattern for a memory bound application. We further observe a performance increase with an increasing number of cores on a single node, but hyper-threading does not further improve the performance. We have used LIKWID [20] to collect bandwidth utilization information and measured a maximum of \(\sim \)140 GB/s using 36 cores (18 per socket) on a single nodeFootnote 9. This matches the achievable bandwidth reported by the STREAM benchmark [11] previously obtained on this machineFootnote 10.
The results in Fig. 7 prove that the performance of \((MP)^{N}\) is bound by memory bandwidth. In each iteration, we access all buffers in a streaming fashion and perform memory operations to keep the underlying buffers with different layout synchronized. We further evaluate the effects of cache-blocking by utilizing smaller tiles, but this approach does not increase the performance due to the lack of reusability and locality in the data structures.
Saturation of performance with an increasing number of MPI processes on a single node with 48 physical cores, \(n=10240\), \(d=128\), \(m=512\). As the structure of the algorithm requires a squared number of processes, we observe speedups up to 36 processes. Starting from 49 processes—the region with the background color of red—hyper-threading causes a performance drop and no additional speedup can be seen. (Color figure online)
Results of strong scaling experiments for computing the matrix profile. We show speedup and parallel efficiency (Top) and a detailed breakdown of time spent in various phases of the application (Bottom). Note that the measurements denoted by \(^{*}\) on the x-axis are executed with exclusive access to the system .
6.4 Scalability
Figure 8 illustrates the speedup and efficiency (top) as well as the runtime decomposition (bottom) for strong scaling experiments. We select a random time series of size \(n=524\,288\), \(d=128\) and \(m=512\), and run these experiments on SuperMUC-NG using 6 to \(1\,366\) nodes (256 cores to \(65\,536\) cores). We observe a linear speedup and throughput of the kernel execution time, using the 6-node configuration as the baseline. However, the time for problem setup—mainly time for creating communicators—increases drastically. This reduces the parallel efficiency to 64% for the experiment with \(1\,366\) nodes.
Similarly, Fig. 9 shows our results for weak scaling experiments. Here, we fix the workload per core \(n_{core}\) to \(2\,048\). This results in a global problem size of \(n=2\,048\) on 1 node with 1 core, scaling up to \(n=1M\) on \(5\,462\) nodes with \(262\,144\) cores. This setup results in weak scaling experiments with kernel execution time of roughly 20 s, which is sufficient for discussions in this paper, and enough to characterize the performance of \((MP)^{N}\) and illustrate dominating overheads. Again, we observe an ideal scaling of kernel execution time. However, we again encounter a significant increase (\(\sim \)58%) in time spent on the creation of communicators. In contrast to the strong scaling results, though, this lies within an acceptable range, as setup time is problem independent and scales only with the number of MPI processes.
Weak scaling of \((MP)^{N}\), illustrating time spent in kernels and other time consuming phases vs. number of cores. We highlight three regions of number of processes: the region with light red color represents the experiments limited to 1 node; the light green shows experiments in middle ranges; and the light blue region represent the experiments that are conducted using more than half of the system resources. Unlike others, the large experiments in blue regions are only done once. (Color figure online)
Even though the setup overheads can be absorbed by the runtime in the more common weak scaling scenario, it clearly represents a performance problem. Further, by comparison with our performance model, we also see that this is not a property of the algorithm, but rather of the system. Upon further investigation, we trace the overhead back to the creation of the column- and row-wise communicators using MPI_Comm_split in the Setup phase when using the default Intel MPI implementation. Subsequent experiments with alternative MPI implementations have shown significantly better scaling for the MPI_Comm_split operation, indicating a non-scalable implementation, which needs to be resolved.
Overall, though, and despite the observed performance problem in MPI, our novel algorithmic approach was able to compute the matrix profile of a 128-dimensional time series dataset of 1 million records on the SuperMUC-NG Petascale system. This corresponds to a projected performance (using measurements from Intel Advisor) of 1.3 Petaflop/s for \((MP)^{N}\) kernels.
6.5 Validation of \((MP)^{N}\)Against Our Model
As can be observed from the presented results in this section (Figs. 7, 8, and 9), Eq. 16 well represents \(T_{Kernel}\), however, this is not the case for Eq. 13, which represents \(T_{Setup}\). We conduct a number of experiments to evaluate the models that we presented in Sect. 5. We execute \((MP)^{N}\) with 283 different combinations of various parameters on the SuperMUC-NG and evaluate \(C_{Setup}\) and \(C_{Kernel}\) to \((8.8 \pm 0.4)\text {e-}06\) and \((5.644 \pm 0.020)\text {e-}09\) respectively. As the suggested analytical model for \(T_{Setup}\) does not properly fit to our experimental data, we used Extra-PFootnote 11 to investigate \(T_{Setup}\), which suggests quadratic growth with the number of MPI processes. This growth corresponds to the performance problem with MPI_Comm_split discussed above.
7 Related Work
There is extensive literature on mining time series, including several survey papers [5, 6, 16]. Esling et al. [5] introduce dimensionality as a fundamental problem in time series mining, and most of the techniques exploit a dimensionality reduction step to address this problem, leaving only limited methods that address similarity search in all dimensions. Matrix profile is superior to alternative approaches like Balasubramanian et al. [1], Tanaka et al. [18] and Vahdatpour et al. [21] in mining similar patterns and provides semantically meaningful multi-dimensional motifs [23].
There are a number of studies on scalable solutions for time series mining: Huang et al. [8] target the use of Apache Spark for speeding discord discovery in time series, however, the conducted experiments in this work are limited to 10 nodes. Berard et al. [2] scale time series similarity search to 20 nodes on a Hadoop cluster. Sart et al. [17] and Zhu et al. [26] use accelerators for speeding time series mining workloads. Movchan et al. [13] study time series similarity search on the Intel Many-core Accelerators, utilizing OpenMP. However, none of the above studies address multi-dimensional datasets, and the analysis is very limited to specific targets of similarity search. Overall, there is a large gap in investigation of scalable multi-dimensional time series mining methods.
Yeh et al. [23,24,25,26] previously developed various algorithms for the calculation of matrix profiles. Their evaluation shows good performance for their solutions in the construction of matrix profiles, but the datasets in the mentioned articles are limited to one-dimensional datasets and/or small problem sizes. The existing work by Yeh et al. [23,24,25,26] is not suited for large dimensionality in such large datasets. To target the high demand of computational power, Zimmerman et al. developed SCAMP [27], a cloud-based framework for the parallel calculation of one-dimensional matrix profiles on multiple GPU-based accelerators. However, the multi-dimensional time series problem is also not covered in their solution.
8 Conclusion
In this work, we present a first scalable solution—\((MP)^{N}\)—for the mining of large-scale multi-dimensional time series targeting CPU-based HPC systems. It comprises optimizations and parallelization of the mSTAMP algorithm. For the first time, this enables the computation of large matrix profiles—as a modern data mining approach—on an HPC system and makes it thereby applicable to large-scale real-world problems. Our parallelization scheme enables scaling up to 256K cores, providing highly scalable throughput and accuracy. With that, we confirm the scalability of the matrix profile approach in mining time series. In our experiments, we performed the fastest and largest (\(1M\times 128\)) multi-dimensional matrix profile ever computed with a projected kernel performance of 1.3 Petaflop/s.
Notes
- 1.
In this case, a record is a collection of samples of all sensors at a specific time.
- 2.
- 3.
Given two distance values a and b, and associated neighboring indices \(i_a\) and \(i_b\), the argmin operation is defined as following: the output (\(i_{out}\)) is set to \(i_a\) if \(a < b\) and is set to \(i_b\) otherwise.
- 4.
Similarity join—also known similarity indexing in the literature—is an operation that combines two input (multi-dimensional) time series and finds similarities. Self-join is the case with two identical input series. For simplicity and without losing generality, we restrict the formulations and discussions to self-joins.
- 5.
Synchronization between the phases is implied by MPI operations used in implementation of \((MP)^{N}\).
- 6.
- 7.
Taxicab norm (\(L_{1}\)) and Infinity norm (\(L_{\infty }\)).
- 8.
For details on mapping MPI processes to cores see https://software.intel.com/en-us/mpi-developer-reference-linux-interoperability-with-openmp.
- 9.
\((MP)^{N}\) requires a quadratic number of MPI processes.
- 10.
We achieved a maximum bandwidth of 185.9 GB/s using STREAM benchmark for copy operation using 48 cores.
- 11.
References
Balasubramanian, A., Wang, J., Prabhakaran, B.: Discovering multidimensional motifs in physiological signals for personalized healthcare. IEEE J. Sel. Topics Signal Process. 10(5), 832–841 (2016). https://doi.org/10.1109/JSTSP.2016.2543679
Berard, A., Hebrail, G.: Searching time series with Hadoop in an electric power company. In: Proceedings of the 2nd International Workshop on Big Data, Streams and Heterogeneous Source Mining: Algorithms, Systems, Programming Models and Applications BigMine 2013, pp. 15–22. ACM, New York (2013). https://doi.org/10.1145/2501221.2501224
Bramas, B.: A novel hybrid Quicksort algorithm vectorized using AVX-512 on Intel Skylake. Int. J. Adv. Comput. Sci. Appl. 8(10) (2017). https://doi.org/10.14569/IJACSA.2017.081044
Chakrabarti, S., et al.: Data mining curriculum: a proposal (version 1.0). In: Intensive Working Group of ACM SIGKDD Curriculum Committee 140 (2006)
Esling, P., Agon, C.: Time-series data mining. ACM Comput. Surv. 45(1), 12:1–12:34 (2012). https://doi.org/10.1145/2379776.2379788
Fu, T.C.: A review on time series data mining. Eng. Appl. Artif. Intell. 24(1), 164–181 (2011). https://doi.org/10.1016/j.engappai.2010.09.007
Gharghabi, S., et al.: Domain agnostic online semantic segmentation for multi-dimensional time series. Data Min. Knowl. Discov. 33, 96–130 (2018)
Huang, T., et al.: Parallel discord discovery. In: Bailey, J., Khan, L., Washio, T., Dobbie, G., Huang, J.Z., Wang, R. (eds.) PAKDD 2016. LNCS (LNAI), vol. 9652, pp. 233–244. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-31750-2_19
Josuttis, N.M.: The C++ Standard Library: A Tutorial and Reference. Addison-Wesley, Boston (2012)
Karlstetter, R., et al.: Turning dynamic sensor measurements from gas turbines into insights: a big data approach. In: Proceedings of the ASME Turbo Expo: Power for Land, Sea, and Air, Volume 6: Ceramics; Controls, Diagnostics, and Instrumentation; Education; Manufacturing Materials and Metallurgy, June 2019. https://doi.org/10.1115/GT2019-91259. v006T05A021
McCalpin, J.D.: Memory bandwidth and machine balance in current high performance computers. IEEE Comput. Soc. Tech. Comm. Comput. Archit. (TCCA) Newsl. 2, 19–25 (1995)
Moody, A., Ahn, D.H., de Supinski, B.R.: Exascale algorithms for generalized MPI\_Comm\_split. In: Cotronis, Y., Danalis, A., Nikolopoulos, D.S., Dongarra, J. (eds.) EuroMPI 2011. LNCS, vol. 6960, pp. 9–18. Springer, Heidelberg (2011). https://doi.org/10.1007/978-3-642-24449-0_4. http://dl.acm.org/citation.cfm?id=2042476.2042480
Movchan, A., Zymbler, M.: Time series subsequence similarity search under dynamic time warping distance on the intel many-core accelerators. In: Amato, G., Connor, R., Falchi, F., Gennaro, C. (eds.) SISAP 2015. LNCS, vol. 9371, pp. 295–306. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-25087-8_28
Netti, A., et al.: From facility to application sensor data: modular, continuous and holistic monitoring with DCDB. In: Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis SC 2019, pp. 64:1–64:27. ACM, New York (2019). https://doi.org/10.1145/3295500.3356191
Pfeilschifter, G.: Time series analysis with matrix profile on HPC systems. Master thesis, Technische Universität München (2019)
Roddick, J.F., Spiliopoulou, M.: A survey of temporal knowledge discovery paradigms and methods. IEEE Trans. Knowl. Data Eng. 14, 750–767 (2002)
Sart, D., Mueen, A., Najjar, W., Keogh, E., Niennattrakul, V.: Accelerating dynamic time warping subsequence search with GPUs and FPGAs. In: 2010 IEEE International Conference on Data Mining, pp. 1001–1006, December 2010
Tanaka, Y., Iwamoto, K., Uehara, K.: Discovery of time-series motif from multi-dimensional data based on MDL principle. Mach. Learn. 58, 269–300 (2005)
Taylor, S.: Optimizing Applications for Multi-Core Processors, Using the Intel Integrated Performance Primitives. Intel Press, Santa Clara (2007)
Treibig, J., Hager, G., Wellein, G.: LIKWID: a lightweight performance-oriented tool suite for x86 multicore environments. In: Proceedings of PSTI2010, the First International Workshop on Parallel Software Tools and Tool Infrastructures, San Diego, CA (2010)
Vahdatpour, A., Amini, N., Sarrafzadeh, M.: Toward unsupervised activity discovery using multi-dimensional motif detection in time series. In: Proceedings of the 21st International Joint Conference on Artifical Intelligence IJCAI 2009, pp. 1261–1266. Morgan Kaufmann Publishers Inc., San Francisco (2009). http://dl.acm.org/citation.cfm?id=1661445.1661647
Yeh, C.M., Herle, H.V., Keogh, E.: Matrix profile III: the matrix profile allows visualization of salient subsequences in massive time series. In: 2016 IEEE 16th International Conference on Data Mining (ICDM), pp. 579–588, December 2016. https://doi.org/10.1109/ICDM.2016.0069
Yeh, C.M., Kavantzas, N., Keogh, E.: Matrix profile VI: meaningful multidimensional motif discovery. In: 2017 IEEE International Conference on Data Mining (ICDM), pp. 565–574, November 2017. https://doi.org/10.1109/ICDM.2017.66
Yeh, C.M., et al.: Matrix profile I: all pairs similarity joins for time series: a unifying view that includes motifs, discords and shapelets. In: 2016 IEEE 16th International Conference on Data Mining (ICDM), pp. 1317–1322, December 2016. https://doi.org/10.1109/ICDM.2016.0179
Zhu, Y., Yeh, C.M., Zimmerman, Z., Kamgar, K., Keogh, E.: Matrix profile XI: SCRIMP++: time series motif discovery at interactive speeds. In: 2018 IEEE International Conference on Data Mining (ICDM), pp. 837–846, November 2018. https://doi.org/10.1109/ICDM.2018.00099
Zhu, Y., et al.: Matrix profile II: exploiting a novel algorithm and GPUs to break the one hundred million barrier for time series motifs and joins. Knowl. Inf. Syst. 54(1), 203–236 (2018)
Zimmerman, Z., et al.: Scaling time series motif discovery with GPUs: breaking the quintillion pairwise comparisons a day barrier. In: Proceedings of the ACM Symposium on Cloud Computing (2018)
Acknowledgments
This work is funded by Bayerische Forschungsstiftung under the research grant Optimierung von Gasturbinen mit Hilfe von Big Data (AZ-1214-16).
The authors gratefully acknowledge the Gauss Centre for Supercomputing e.V. (https://www.gauss-centre.eu) for funding this work by providing computing time on the GCS Supercomputer SuperMUC-NG at Leibniz Supercomputing Centre (LRZ). Also, we would also like to thank LRZ’s staff for their valuable support.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Raoofy, A., Karlstetter, R., Yang, D., Trinitis, C., Schulz, M. (2020). Time Series Mining at Petascale Performance. In: Sadayappan, P., Chamberlain, B.L., Juckeland, G., Ltaief, H. (eds) High Performance Computing. ISC High Performance 2020. Lecture Notes in Computer Science(), vol 12151. Springer, Cham. https://doi.org/10.1007/978-3-030-50743-5_6
Download citation
DOI: https://doi.org/10.1007/978-3-030-50743-5_6
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-50742-8
Online ISBN: 978-3-030-50743-5
eBook Packages: Computer ScienceComputer Science (R0)