Abstract
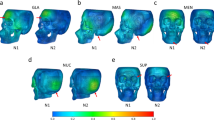
We train a convolutional neural network (CNN) to classify the sex of mice from x-ray images in order to develop a tool that can be used not only for quality control of high throughput image data, but also to identify sex-related phenotype alterations. The method achieved 98% accuracy, recall of .98 and precision of .98 and identified the chest and pelvis as the areas relevant for sex classification. We identified four knockout lines (Duoxa2, Tmem189, Dusp3 and Il10rb) potentially affected by phenotype changes related to sex.
This study demonstrates that CNNs can be trained for the purpose of quality control of images and can aid the discovery of novel genotype-phenotype associations. In addition to facilitating quality control, the method presented (1) allows the creation of a tool that will help phenotypers flag images of mice that should be inspected in more detail, (2) has highlighted areas of the mouse that are of particular interest in phenotype changes related to sex, and (3) has the potential to identify genes that may be causing sex related phenotype changes and/or are involved in sexual dimorphism.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Dickinson, M., Flenniken, A., Ji, X., et al.: High-throughput discovery of novel developmental phenotypes. Nature 537, 508–514 (2016). https://doi.org/10.1038/nature19356
LeCun, Y., Bengio, Y., Hinton, G.: Deep learning. Nature 521, 436–444 (2015)
Litjens, G., et al.: A survey on deep learning in medical image analysis. Med. Image Anal. 42, 60–88 (2017)
Paszke, A., Gross, S., et al.: PyTorch: an imperative style, high-performance deep learning library. In: Wallach, H., Larochelle, H., Beygelzimer, A., d’ Alche-Buc, F., Fox, E., Garnett, R. (eds.) Advances in Neural Information Processing Systems 32, pp. 8024–8035. Curran Associates, Inc. (2019). http://papers.neurips.cc/paper/9015-pytorch-an-imperative-style-high-performance-deep-learning-library.pdf
Simonyan, K., Zisserman, A.: Very deep convolutional networks for large-scale image recognition. In: 3rd International Conference on Learning Representations, ICLR 2015, San Diego, CA, USA, 7–9 May 2015, Conference Track Proceedings (2015). http://arxiv.org/abs/1409.1556
Vandereyken, M.M., et al.: Dual-specificity phosphatase 3 deletion protects female, but not male, mice from endotoxemia-induced andpolymicrobial-induced septic shock. J. Immunol. 199(7), 2525–2527 (2017). https://doi.org/10.4049/jimmunol.1602092
Yosinski, J., Clune, J., Bengio, Y., Lipson, H.: How transferable are features in deep neural networks? In: Ghahramani, Z., Welling, M., Cortes, C., Lawrence, N.D., Weinberger, K.Q. (eds.) Advances in Neural Information Processing Systems 27, pp. 3320–3328 (2014). http://papers.nips.cc/paper/5347-how-transferable-are-features-in-deep-neural-networks.pdf
Zeiler, M.D., Fergus, R.: Visualizing and understanding convolutional networks. In: Fleet, D., Pajdla, T., Schiele, B., Tuytelaars, T. (eds.) ECCV 2014. LNCS, vol. 8689, pp. 818–833. Springer, Cham (2014). https://doi.org/10.1007/978-3-319-10590-1_53
Zhou, B., Khosla, A., Lapedriza, A., Oliva, A., Torralba, A.: Learning deep features for discriminative localization. In: CVPR (2016)
Acknowledgements
Our thanks to Professor Jesús Ruberte of UAB-Barcelona, and Dr David Lafont of the Wellcome Trust Sanger Institute for insightful discussions about the physiological relevance of the CAMs. This work was supported by the United States National Institutes of Health (NIH) Grant U54 HG006370.
Author information
Authors and Affiliations
Consortia
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Babalola, K., Mashhadi, H.H., Muñoz-Fuentes, V., Mason, J., Meehan, T., On Behalf of the International Mouse Phenotyping Consortium. (2020). Investigating Sex Related Phenotype Changes in Knockout Mice by Applying Deep Learning to X-Ray Images. In: Papież, B., Namburete, A., Yaqub, M., Noble, J. (eds) Medical Image Understanding and Analysis. MIUA 2020. Communications in Computer and Information Science, vol 1248. Springer, Cham. https://doi.org/10.1007/978-3-030-52791-4_28
Download citation
DOI: https://doi.org/10.1007/978-3-030-52791-4_28
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-52790-7
Online ISBN: 978-3-030-52791-4
eBook Packages: Computer ScienceComputer Science (R0)