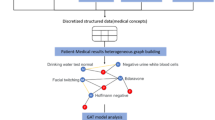
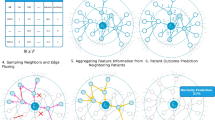
Abstract
Electronic Health Record (EHR) data is a rich source for powerful biomedical discovery but it consists of a wide variety of data types that are traditionally difficult to model. Furthermore, many machine learning frameworks that utilize these data for predictive tasks do not fully leverage the inter-connectivity structure and therefore may not be fully optimized. In this work, we propose a relational, deep heterogeneous network learning method that operates on EHR data and addresses these limitations. In this model, we used three different node types: patient, lab, and diagnosis. We show that relational graph learning naturally encodes structured relationships in the EHR and outperforms traditional multilayer perceptron models in the prediction of thousands of diseases. We evaluated our model on EHR data derived from MIMIC-III, a public critical care data set, and show that our model has improved prediction of numerous disease diagnoses.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Bordes, A., Usunier, N., Garcia-Duran, A., Weston, J., Yakhnenko, O.: Translating embeddings for modeling multi-relational data. In: Advances in Neural Information Processing Systems, pp. 2787–2795 (2013)
Choi, E., Bahadori, M.T., Schuetz, A., Stewart, W.F., Sun, J.: Doctor AI: predicting clinical events via recurrent neural networks. In: Machine Learning for Healthcare Conference, pp. 301–318 (2016)
Choi, E., Xiao, C., Stewart, W., Sun, J.: Mime: multilevel medical embedding of electronic health records for predictive healthcare. In: Advances in Neural Information Processing Systems, pp. 4547–4557 (2018)
Choi, E., et al.: Graph convolutional transformer: learning the graphical structure of electronic health records. arXiv preprint arXiv:1906.04716 (2019)
Dong, Y., Chawla, N.V., Swami, A.: metapath2vec: scalable representation learning for heterogeneous networks. In: Proceedings of the 23rd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp. 135–144 (2017)
Glicksberg, B.S., Johnson, K.W., Dudley, J.T.: The next generation of precision medicine: observational studies, electronic health records, biobanks and continuous monitoring. Hum. Mol. Genet. 27(R1), R56–R62 (2018)
Glicksberg, B.S., et al.: Automated disease cohort selection using word embeddings from electronic health records. In: World Scientific (2018)
Lipton, Z.C., Kale, D.C., Elkan, C., Wetzel, R.: Learning to diagnose with LSTM recurrent neural networks. arXiv preprint arXiv:1511.03677 (2015)
Liu, C., Wang, F., Hu, J., Xiong, H.: Temporal phenotyping from longitudinal electronic health records: a graph based framework. In: Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp. 705–714 (2015)
Malinchoc, M., Kamath, P.S., Gordon, F.D., Peine, C.J., Rank, J., Ter Borg, P.C.: A model to predict poor survival in patients undergoing transjugular intrahepatic portosystemic shunts. Hepatology 31(4), 864–871 (2000)
Mikolov, T., Sutskever, I., Chen, K., Corrado, G.S., Dean, J.: Distributed representations of words and phrases and their compositionality. In: Advances in Neural Information Processing Systems, pp. 3111–3119 (2013)
Miotto, R., Li, L., Kidd, B.A., Dudley, J.T.: Deep patient: an unsupervised representation to predict the future of patients from the electronic health records. Sci. Rep. 6(1), 1–10 (2016)
Shickel, B., Tighe, P.J., Bihorac, A., Rashidi, P.: Deep EHR: a survey of recent advances in deep learning techniques for electronic health record (EHR) analysis. IEEE J. Biomed. Health Inform. 22(5), 1589–1604 (2017)
Zhu, Z., Yin, C., Qian, B., Cheng, Y., Wei, J., Wang, F.: Measuring patient similarities via a deep architecture with medical concept embedding. In: 2016 IEEE 16th International Conference on Data Mining (ICDM), pp. 749–758. IEEE (2016)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
A Appendix
A Appendix
Definition 1 (Heterogeneous Network)
A heterogeneous network is defined as a graph \(\mathbf{G} =(V,E,T)\), where each node \(\mathbf{v} \) and each link \(\mathbf{e} \) are represented by their mapping functions to a specific node and relation type \(\phi (v):V\rightarrow T_{V}\) and \(\phi (e):E\rightarrow T_{E}\). Where \(T_{V}\) and \(T_{E}\) denote the sets of node and relation types, and \(|T_{V}|+|T_{E}|>2\).
Definition 2 (Heterogeneous Graph Learning)
Given a heterogeneous network \(\mathbf{G} \), the task of heterogeneous graph learning is to learn a function mapping \(f:V\rightarrow {R^{d}}\), that connects disparate type of nodes into a \(d-dimensional\) uniform latent representation \(X\in R^{|V|\times d}\), and \(d\ll |V|\), that are able to capture the structural and semantic relations between them.
Definition 3 (One-hop Connectivity)
One-hop connectivity in a heterogeneous network is the local pairwise connection between two consecutive vertices, which directly linked by an edge belongs to a relational type.
1.1 A.1 Skip-Gram Model
The skip-gram model [11] seeks to maximize the probability of observing the context neighborhood nodes given the center node:
Where \(N_c(u)\) is the neighborhood context nodes of the center node u, and f(u) is the latent representation of u.
1.2 A.2 Heterogeneous Skip-Gram Model
EHR data is heterogeneous, including varies type of vertices, such as lab tests, diagnoses, prescriptions, and patient demographics. Each of these vertices encodes different information. Heterogeneous Skip-gram model [5] learns the latent expression of these different type of nodes by maximizing the probability of observing heterogeneous neighborhood given a center node:
Where \(N_{t}(u)\) is the heterogeneous neighborhood vertices of center node u, and \(t\in T_{V}\) is the node type.
1.3 A.3 TransE
TransE model [1] aims to relate different type of nodes by their relationship type. Specifically, two different types of nodes are connected by a relation type would be represented as a triple (head, relation, tail), denoted as (h, l, t). For example, one triple from EHR data could be (patient, diagnosed, ICD), where patient is the head node, ICD is the specific diagnosis code attributed to the patient, and the relation between these two vertices is diagnosed.
This TransE model leverages the procedure by first projecting different type of node with different initial representation dimension into a same latent dimension space (where the dimension of this latent space can be customized), and these two different type projected nodes are linked by a relation type which is represented as a translation vector in that latent space. Both the projection matrix and the relational translation vector are learnable parameters in the deep learning system.
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Wanyan, T. et al. (2020). Heterogeneous Graph Embeddings of Electronic Health Records Improve Critical Care Disease Predictions. In: Michalowski, M., Moskovitch, R. (eds) Artificial Intelligence in Medicine. AIME 2020. Lecture Notes in Computer Science(), vol 12299. Springer, Cham. https://doi.org/10.1007/978-3-030-59137-3_2
Download citation
DOI: https://doi.org/10.1007/978-3-030-59137-3_2
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-59136-6
Online ISBN: 978-3-030-59137-3
eBook Packages: Computer ScienceComputer Science (R0)