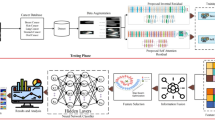
Abstract
Medical image analysis benefits Computer Aided Diagnosis (CADx). A fundamental analyzing approach is the classification of medical images, which serves for skin lesion diagnosis, diabetic retinopathy grading, and cancer classification on histological images. When learning these discriminative classifiers, we observe that the convolutional neural networks (CNNs) are vulnerable to distractor interference. This is due to the similar sample appearances from different categories (i.e., small inter-class distance). Existing attempts select distractors from input images by empirically estimating their potential effects to the classifier. The essences of how these distractors affect CNN classification are not known. In this paper, we explore distractors from the CNN feature space via proposing a neuron intrinsic learning method. We formulate a novel distractor-aware loss that encourages large distance between the original image and its distractor in the feature space. The novel loss is combined with the original classification loss to update network parameters by back-propagation. Neuron intrinsic learning first explores distractors crucial to the deep classifier and then uses them to robustify CNN inherently. Extensive experiments on medical image benchmark datasets indicate that the proposed method performs favorably against the state-of-the-art approaches.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
APTOS 2019 Blindness Detection (2019). https://www.kaggle.com/c/aptos2019-blindness-detection/data
Deng, J., Dong, W., Socher, R., Li, L.J., Li, K., Fei-Fei, L.: Imagenet: a large-scale hierarchical image database. In: IEEE Conference on Computer Vision and Pattern Recognition, pp. 248–255. IEEE (2009)
Haofu, L., Luo, J.: A deep multi-task learning approach to skin lesion classification. In: Workshops at the Thirty-First AAAI Conference on Artificial Intelligence (2017)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
Kather, J.N.: Histological images for MSI vs. MSS classification in gastrointestinal cancer. FFPE Samples (2019). https://doi.org/10.5281/zenodo.2530835
Li, X., Hu, X., Yu, L., Zhu, L., Fu, C.W., Heng, P.A.: Canet: cross-disease attention network for joint diabetic retinopathy and diabetic macular edema grading. IEEE Trans. Med. Imaging 39, 1483–1493 (2019)
Litjens, G., et al.: A survey on deep learning in medical image analysis. Med. Image Anal. 42, 60–88 (2017)
Loshchilov, I., Hutter, F.: Online batch selection for faster training of neural networks (2015). arXiv preprint arXiv:1511.06343
Rakhlin, A.: Diabetic retinopathy detection through integration of deep learning classification framework. bioRxiv, p. 225508 (2018)
Shrivastava, A., Gupta, A., Girshick, R.: Training region-based object detectors with online hard example mining. In: IEEE Conference on Computer Vision and Pattern Recognition, pp. 761–769 (2016)
Simonyan, K., Vedaldi, A., Zisserman, A.: Deep inside convolutional networks: Visualising image classification models and saliency maps (2013). arXiv preprint arXiv:1312.6034
Simonyan, K., Zisserman, A.: Very deep convolutional networks for large-scale image recognition (2014). arXiv preprint arXiv:1409.1556
Song, Y., Bao, L., He, S., Yang, Q., Yang, M.H.: Stylizing face images via multiple exemplars. Comput. Vis. Image Underst. 162, 135–145 (2017)
Song, Y., et al.: Joint face hallucination and deblurring via structure generation and detail enhancement. Int. J. Comput. Vis. 127, 785–800 (2019)
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., Wojna, Z.: Rethinking the inception architecture for computer vision. In: IEEE Conference on Computer Vision and Pattern Recognition, pp. 2818–2826 (2016)
Tajbakhsh, N., et al.: Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans. Med. Imaging 35(5), 1299–1312 (2016)
Tan, M., Le, Q.V.: Efficientnet: Rethinking model scaling for convolutional neural networks (2019). arXiv preprint arXiv:1905.11946
Tschandl, P., Rosendahl, C., Kittler, H.: The ham10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 5, 180161 (2018)
Wang, W., et al.: Medical image classification using deep learning. In: Chen, Y.-W., Jain, L.C. (eds.) Deep Learning in Healthcare. ISRL, vol. 171, pp. 33–51. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-32606-7_3
Zhu, Z., Wang, Q., Li, B., Wu, W., Yan, J., Hu, W.: Distractor-aware siamese networks for visual object tracking. In: European Conference on Computer Vision, pp. 101–117 (2018)
Acknowledgments
This work was funded by the Key Area Research and Development Program of Guangdong Province, China (No. 2018B010111001), National Key Research and Development Project (2018YFC2000702) and Science and Technology Program of Shenzhen, China (No. ZDSYS201802021814180).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Gong, L., Ma, K., Zheng, Y. (2020). Distractor-Aware Neuron Intrinsic Learning for Generic 2D Medical Image Classifications. In: Martel, A.L., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2020. MICCAI 2020. Lecture Notes in Computer Science(), vol 12262. Springer, Cham. https://doi.org/10.1007/978-3-030-59713-9_57
Download citation
DOI: https://doi.org/10.1007/978-3-030-59713-9_57
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-59712-2
Online ISBN: 978-3-030-59713-9
eBook Packages: Computer ScienceComputer Science (R0)