Abstract
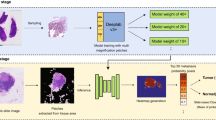
Segmenting histology images is challenging because of the sheer size of the images with millions or even billions of pixels. Typical solutions pre-process each histology image by dividing it into patches of fixed size and/or down-sampling to meet memory constraints. Such operations incur information loss in the field-of-view (FoV) (i.e., spatial coverage) and the image resolution. The impact on segmentation performance is, however, as yet understudied. In this work, we first show under typical memory constraints (e.g., 10G GPU memory) that the trade-off between FoV and resolution considerably affects segmentation performance on histology images, and its influence also varies spatially according to local patterns in different areas (see Fig. 1). Based on this insight, we then introduce foveation module, a learnable “dataloader” which, for a given histology image, adaptively chooses the appropriate configuration (FoV/resolution trade-off) of the input patch to feed to the downstream segmentation model at each spatial location (Fig. 1). The foveation module is jointly trained with the segmentation network to maximise the task performance. We demonstrate, on the Gleason2019 challenge dataset for histopathology segmentation, that the foveation module improves segmentation performance over the cases trained with patches of fixed FoV/resolution trade-off. Moreover, our model achieves better segmentation accuracy for the two most clinically important and ambiguous classes (Gleason Grade 3 and 4) than the top performers in the challenge by 13.1% and 7.5%, and improves on the average performance of 6 human experts by 6.5% and 7.5%.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Notes
- 1.
Screen display or human vision typically have lower resolutions than that of the ultra-high resolution images of interest in this work.
- 2.
References
Srinidhi, C.L., Ciga, O., Martel, A.L.: Deep neural network models for computational histopathology: asurvey. arXiv preprint arXiv:1912.12378 (2019)
Seth, N., Akbar, S., Nofech-Mozes, S., Salama, S., Martel, A.L.: Automated segmentation of DCIS in whole slide images. In: Reyes-Aldasoro, C.C., Janowczyk, A., Veta, M., Bankhead, P., Sirinukunwattana, K. (eds.) ECDP 2019. LNCS, vol. 11435, pp. 67–74. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-23937-4_8
Kamnitsas, K., et al.: Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 36, 61–78 (2017)
Chen, L.-C., Yang, Y., Wang, J., Xu, W., Yuille, A.L.: Attention to scale: scale-aware semantic image segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 3640–3649 (2016)
Chen, W., Jiang, Z., Wang, Z., Cui, K., Qian, X.: Collaborative global-local networks for memory-efficient segmentation of ultra-high resolution images. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 8924–8933 (2019)
Li, Y., Junmin, W., Qisong, W.: Classification of breast cancer histology images using multi-size and discriminative patches based on deep learning. IEEE Access 7, 21400–21408 (2019)
Katharopoulos, A., Fleuret, F.: Processing megapixel images with deep attention-sampling models. arXiv preprint arXiv:1905.03711 (2019)
Warfield, S.K., Zou, K.H., Wells, W.M.: Simultaneous truth and performance level estimation (staple): an algorithm for the validation of image segmentation. IEEE Trans. Med. Imaging 23(7), 903–921 (2004)
Ilse, M., Tomczak, J.M., Welling, M.: Attention-based deep multiple instance learning. arXiv preprint arXiv:1802.04712 (2018)
Xu, K., et al.: Show, attend and tell: Neural image caption generation with visual attention. In: International Conference on Machine Learning, pp. 2048–2057 (2015)
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980 (2014)
He, K., Zhang, X., Ren, S., Sun, J.: Delving deep into rectifiers: surpassing human-level performance on imagenet classification. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 1026–1034 (2015)
Sun, K., et al.: High-resolution representations for labeling pixels and regions. arXiv preprint arXiv:1904.04514 (2019)
Acknowledgements
We sincerely acknowledge: Marnix Jansen for inspirational pathological advice. Hongxiang Lin for the insightful discussions. C.J., T.M. and D.A. acknowledge funding by the EPSRC grants EP/R006032/1, EP/M020533/1, the CRUK/EPSRC grant NS/A000069/1, and the NIHR UCLH Biomedical Research Centre.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
1 Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 2 (mp4 143 KB)
Supplementary material 3 (mp4 186 KB)
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Jin, C., Tanno, R., Xu, M., Mertzanidou, T., Alexander, D.C. (2020). Foveation for Segmentation of Mega-Pixel Histology Images. In: Martel, A.L., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2020. MICCAI 2020. Lecture Notes in Computer Science(), vol 12265. Springer, Cham. https://doi.org/10.1007/978-3-030-59722-1_54
Download citation
DOI: https://doi.org/10.1007/978-3-030-59722-1_54
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-59721-4
Online ISBN: 978-3-030-59722-1
eBook Packages: Computer ScienceComputer Science (R0)