Abstract
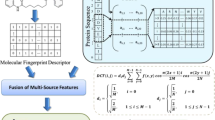
Drug molecules interact with target proteins to influence the pharmacological action of the target to achieve the phenotypic effect, which can facilitate the identification of novel targets for current drug. Traditional biological experiments for discovering new drug-target interactions are expensive and time-consuming. Therefore, it is crucial to develop new prediction methods for identifying potential drug-target interactions. Computing methods have been increasing developed which can quickly and effectively predict drug-target interactions. In particular, machine learning methods have been widely used due to high predictive performance and computational efficiency. This paper first uses MACCS substructure fingerings to encode the drug molecules, then uses CNNs to extract the biological evolutionary information of target protein sequences, and finally uses random forest algorithm to predict drug-target interactions. Four datasets of drug-target interactions including Enzymes, Ion Channels, GPCRs and Nuclear Receptors, are independently used for building models with random forest. The results demonstrate our proposed method has a general compatibility, which is effective and feasible to predict drug-target interactions.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Kim, I.W., Jang, H., Hyunkim, J., et al.: Computational drug repositioning for gastric cancer using reversal gene expression profiles. Sci. Rep. 9, 2660 (2019)
Ganotra, G.K., Wade, R.C.: Prediction of drug-target binding kinetics by comparative binding energy analysis. ACS Med. Chem. Lett. 9(11), 1134–1139 (2018)
Kingsmore, K.M., Grammer, A.C., Lipsky, P.E.: Drug repurposing to improve treatment of rheumatic autoimmune inflammatory diseases. Nat. Rev. Rheumatol. 16, 32–52 (2020)
Williams, G., Gatt, A., Clarke, E., et al.: Drug repurposing for Alzheimer’s disease based on transcriptional profiling of human iPSC-derived cortical neurons. Transl. Psychiatry 9, 220 (2019)
Stokes, J.M., Yang, K., Swanson, K., et al.: A deep learning approach to antibiotic discovery. Cell 180(4), 668–702 (2020)
Zhang, W., Lin, W., Zhang, D., Wang, S., Shi, J., Niu, Y.: Recent advances in the machine learning-based drug-target interaction prediction. Curr. Drug Metab. 20(3), 194–202 (2019)
Peng, L., Liao, B., Zhu, W., Li, Z., Li, K.: Predicting drug-target interactions with multi-information fusion. IEEE J Biomed. Health Inf. 21(2), 56–72 (2017)
Zong, N., Kim, H., Ngo, V., Harismendy, O.: Deep mining heterogeneous networks of biomedical linked data to predict novel drug-target associations. Bioinformatics 33(15), 2337–2344 (2017)
Wen, M., et al.: Deep-learning-based drug-target interaction prediction. J. Proteome 16(4), 1401–1409 (2017)
Pliakos, K., Vens, C., Tsoumakas, G.: Predicting drug-target interactions with multi-label classification and label partitioning. IEEE/ACM Trans. Comput. Biol. Bioinf. (2019, early access). https://doi.org/10.1109/TCBB.2019.2951378. https://ieeexplore.ieee.org/document/8890853
Pliakos, K.: Mining biomedical networks exploiting structure and background information. KU Leuven, Belgium (2019)
Zhang, W., Chen, Y., Li, D.: Drug-target interaction prediction through label propagation with linear neighborhood information. Molecules 22(12), 2056 (2017)
Ezzat, A., Zhao, P., Wu, M., Li, X., Kwoh, C.: Drug-target interaction prediction with graph regularized matrix factorization. IEEE/ACM Trans. Comput. Biol. Bioinf. 14(3), 646–656 (2017)
Olayan, R.S., Ashoor, H., Bajic, V.B.: DDR: efficient computational method to predict drug-target interactions using graph mining and machine learning approaches. Bioinformatics 34(7), 1164–1173 (2017)
Li, Z., et al.: Identification of drug-target interaction from interactome network with ‘guilt-by-association’ principle and topology features. Bioinformatics 32(7), 1057–1064 (2016)
Rayhan, F., Ahmed, S., Shatabda, S., et al.: iDTI-ESBoost: identification of drug target interaction using evolutionary and structural features with boosting. Sci. Rep. 7(1), 17731 (2017)
Mousavian, Z., Khakabimamaghani, S., Kavousi, K., et al.: Drug-target interaction prediction from PSSM based evolutionary information. J. Pharmacol. Toxicol. Methods 78, 42–51 (2016)
Öztürk, H., Özgür, A., Ozkirimli, E.: DeepDTA: deep drug-target binding affinity prediction. Bioinformatics 34(17), i821–i829 (2018)
Davis, M.I., Hunt, J.P., Herrgard, S., et al.: Comprehensive analysis of kinase inhibitor selectivity. Nat. Biotechnol. 29(11), 1046–1051 (2011)
He, T., Heidemeyer, M., Ban, F., et al.: SimBoost: a read-across approach for predicting drug-target binding affinities using gradient boosting machines. J. Cheminf. 9(1), 24 (2017)
Tang, J., Szwajda, A., Shakyawar, S., et al.: Making sense of large-scale kinase inhibitor bioactivity data sets: a comparative and integrative analysis. J. Chem. Inf. Model. 54(3), 735–743 (2014)
Yamanishi, Y., Araki, M., Gutteridge, A., et al.: Prediction of drug-target interaction networks from the integration of chemical and genomic spaces. Intell. Syst. Mol. Biol. 24(13), 232–240 (2008)
Yamanishi, Y., Masaaki, K., Minoru, K., et al.: Drug-target interaction prediction from chemical, genomic and pharmacological data in an integrated framework. Bioinformatics 26(12), 246–254 (2010)
Cao, D., Liu, S., Xu, Q., et al.: Large-scale prediction of drug-target interactions using protein sequences and drug topological structures. Anal. Chim. Acta 752, 1–10 (2012)
Gunther, S., Kuhn, M., Dunkel, M., et al.: SuperTarget and matador: resources for exploring drug-target relationships. Nucleic Acids Res. 36, 919–922 (2007)
Kanehisa, M., Goto, S., Hattori, M., et al.: From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 34(90001), 354–357 (2006)
Wishart, D.S., Knox, C., Guo, A.C., et al.: Drugbank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 36(suppl 1), D901–D906 (2008)
Jeske, L., Placzek, S., Schomburg, I., et al.: BRENDA in 2019: a European ELIXIR core data resource. Nucleic Acids Res. 47, 542–549 (2019)
Cheng, F., Liu, C., Jiang, J., et al.: Prediction of drug-target interactions and drug repositioning via network-based inference. PLoS Comput. Biol. 8(5), e1002503 (2012)
Chen, H., Zhang, Z.: A semi-supervised method for drug-target interaction prediction with consistency in networks. PLoS ONE 8(5), e62975 (2013)
Acknowledgment
The authors thank the members of Machine Learning and Artificial Intelligence Laboratory, School of Computer Science and Technology, Wuhan University of Science and Technology, for their helpful discussion within seminars. This work was supported in part by Hubei Province Natural Science Foundation of China (No. 2018CFB526, 2019CFB797), by National Natural Science Foundation of China (No. 61502356, 61972299, 61702385).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Lin, X., Xu, M., Yu, H. (2020). Prediction of Drug-Target Interactions with CNNs and Random Forest. In: Huang, DS., Jo, KH. (eds) Intelligent Computing Theories and Application. ICIC 2020. Lecture Notes in Computer Science(), vol 12464. Springer, Cham. https://doi.org/10.1007/978-3-030-60802-6_32
Download citation
DOI: https://doi.org/10.1007/978-3-030-60802-6_32
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-60801-9
Online ISBN: 978-3-030-60802-6
eBook Packages: Computer ScienceComputer Science (R0)