Abstract
Due to the huge amount of available omic data, classifying samples according to various omics is a complex process. One of the most common approaches consists of creating a classifier for each omic and subsequently making a consensus among the classifiers that assigns to each sample the most voted class among the outputs on the individual omics.
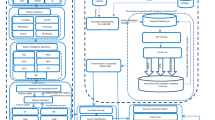
However, this approach does not consider the confidence in the prediction ignoring that a biological information coming from a certain omic may be more reliable than others. Therefore, it is here proposed a method consisting of a tree-based multi-layer perceptron (MLP), which estimates the class-membership probabilities for classification. In this way, it is not only possible to give relevance to all the omics, but also to label as Unknown those samples for which the classifier is uncertain in its prediction. The method was applied to a dataset composed of 909 kidney cancer samples for which these three omics were available: gene expression (mRNA), microRNA expression (miRNA) and methylation profiles (meth) data. The method is valid also for other tissues and on other omics (e.g. proteomics, copy number alterations data, single nucleotide polymorphism data). The accuracy and weighted average f1-score of the model are both higher than 95%. This tool can therefore be particularly useful in clinical practice, allowing physicians to focus on the most interesting and challenging samples.
Data availability: the code is freely accessible at https://github.com/Bontempogianpaolo1/Consunsus-on-multi-omics, while mRNA, miRNA and meth data can be obtained from the GDC database [2] or upon request to the authors.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Weinstein, J.N., et al.: The cancer genome atlas pan-cancer analysis project. Nat. Genet. 45(10), 1113 (2013)
Grossman, R.L., et al.: Toward a shared vision for cancer genomic data. N. Engl. J. Med. 375(12), 1109–1112 (2016)
Leinonen, R., Sugawara, H., Shumway, M.: International nucleotide sequence database collaboration: the sequence read archive. Nucleic Acids Res. 39((suppl_1)), D19–D21 (2010)
Pochet, N., De Smet, F., Suykens, J.A., De Moor, B.L.: Systematic benchmarking of microarray data classification: assessing the role of non-linearity and dimensionality reduction. Bioinformatics 20(17), 3185–3195 (2004)
Lee, G., Rodriguez, C., Madabhushi, A.: An empirical comparison of dimensionality reduction methods for classifying gene and protein expression datasets. In: Măndoiu, I., Zelikovsky, A. (eds.) ISBRA 2007. LNCS, vol. 4463, pp. 170–181. Springer, Heidelberg (2007). https://doi.org/10.1007/978-3-540-72031-7_16
Kim, P.M., Tidor, B.: Subsystem identification through dimensionality reduction of large-scale gene expression data. Genome Res. 13(7), 1706–1718 (2003)
Lu, M., Zhan, X.: The crucial role of multiomic approach in cancer research and clinically relevant outcomes. EPMA J. 9(1), 77–102 (2018)
Wang, B., Mezlini, A.M., Demir, F., Fiume, M., Tu, Z., Brudno, M., et al.: Similarity network fusion for aggregating data types on a genomic scale. Nat. Meth. 11(3), 333 (2014)
Argelaguet, R., et al.: Multi-omics factor analysis—a framework for unsupervised integration of multi-omics data sets. Mol. Syst. Biol. 14(6), e8124 (2018)
Robles, A.I., Arai, E., Mathé, E.A., Okayama, H., Schetter, A.J., Brown, D., et al.: An integrated prognostic classifier for stage I lung adenocarcinoma based on mRNA, microRNA, and DNA methylation biomarkers. J. Thorac. Oncol. 10(7), 1037–1048 (2015)
Tang, W., Wan, S., Yang, Z., Teschendorff, A.E., Zou, Q.: Tumor origin detection with tissue-specific miRNA and DNA methylation markers. Bioinformatics 34(3), 398–406 (2018)
Cantini, L., Medico, E., Fortunato, S., Caselle, M.: Detection of gene communities in multi-networks reveals cancer drivers. Sci. Rep. 5, 17386 (2015)
Barabási, A.L., Gulbahce, N., Loscalzo, J.: Network medicine: a network-based approach to human disease. Nat. Rev. Genet. 12(1), 56–68 (2011)
Fuchs, M., Beißbarth, T., Wingender, E., Jung, K.: Connecting high-dimensional mRNA and miRNA expression data for binary medical classification problems. Comput. Meth. Programs Biomed. 111(3), 592–601 (2013)
Mallik, S., Mukhopadhyay, A., Maulik, U., Bandyopadhyay, S.: Integrated analysis of gene expression and genome-wide DNA methylation for tumor prediction: an association rule mining-based approach. In: 2013 IEEE Symposium on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), (pp. 120–127). IEEE April 2013
Huber, W., Von Heydebreck, A., Sültmann, H., Poustka, A., Vingron, M.: Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18((suppl_1)), S96–S104 (2002)
Love, M.I., Huber, W., Anders, S.: Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15(12), 550 (2014)
Wold, S., Esbensen, K., Geladi, P.: Principal component analysis. Chemometr. Intell. Lab. Syst. 2(1-3), 37–52 (1987)
Breiman, L.: Random forests. Mach. Learn. 45(1), 5–32 (2001)
Ramchoun, H., Idrissi, M.A.J., Ghanou, Y., Ettaouil, M.: Multilayer perceptron: architecture optimization and training. IJIMAI 4(1), 26–30 (2016)
Christopher, M.: Bishop.: Pattern Recognition and Machine Learning (Information Science and Statistics). Springer-Verlag, Berlin, Heidelberg (2006)
Paszke, A., et al. Automatic differentiation in pytorch (2017)
Bingham, E., et al.: Pyro: deep universal probabilistic programming. J. Mach. Learn. Res. 20(1), 973–978 (2019)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Lovino, M., Bontempo, G., Cirrincione, G., Ficarra, E. (2020). Multi-omics Classification on Kidney Samples Exploiting Uncertainty-Aware Models. In: Huang, DS., Jo, KH. (eds) Intelligent Computing Theories and Application. ICIC 2020. Lecture Notes in Computer Science(), vol 12464. Springer, Cham. https://doi.org/10.1007/978-3-030-60802-6_4
Download citation
DOI: https://doi.org/10.1007/978-3-030-60802-6_4
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-60801-9
Online ISBN: 978-3-030-60802-6
eBook Packages: Computer ScienceComputer Science (R0)