Abstract
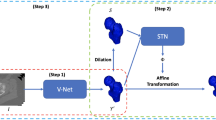
Alignment of contrast and non contrast-enhanced imaging is essential for quantification of changes in several biomedical applications. In particular, the extraction of cartilage shape from contrast-enhanced Computed Tomography (CT) of tibiae requires accurate alignment of the bone, currently performed manually. Existing deep learning-based methods for alignment require a common template or are limited in rotation range. Therefore, we present a novel network, D-net, to estimate arbitrary rotation and translation between 3D CT scans that additionally does not require a prior template. D-net is an extension to the branched Siamese encoder-decoder structure connected by new mutual, non-local links, which efficiently capture long-range connections of similar features between two branches. The 3D supervised network is trained and validated using preclinical CT scans of mouse tibiae with and without contrast enhancement in cartilage. The presented results show a significant improvement in the estimation of CT alignment, outperforming the current comparable methods.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others

References
Baiker, M., Staring, M., Löwik, C.W.G.M., Reiber, J.H.C., Lelieveldt, B.P.F.: Automated registration of whole-body follow-up MicroCT data of mice. In: Fichtinger, G., Martel, A., Peters, T. (eds.) MICCAI 2011. LNCS, vol. 6892, pp. 516–523. Springer, Heidelberg (2011). https://doi.org/10.1007/978-3-642-23629-7_63
Chee, E., Wu, Z.: Airnet: self-supervised affine registration for 3d medical images using neural networks. arXiv preprint arXiv:1810.02583 (2018)
Dunnhofer, M., et al.: Siam-U-Net: encoder-decoder siamese network for knee cartilage tracking in ultrasound images. Med. Image Anal. 60, 101631 (2020)
Haskins, G., et al.: Learning deep similarity metric for 3D MR-TRUS image registration. Int. J. Comput. Assist. Radiol. Surg. 14(3), 417–425 (2019)
Haskins, G., Kruger, U., Yan, P.: Deep learning in medical image registration: a survey. Mach. Vis. Appl. 31(1), 8 (2020)
Hu, Y., et al.: Label-driven weakly-supervised learning for multimodal deformable image registration. In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), pp. 1070–1074. IEEE (2018)
Kwon, D., et al.: Siamese U-net with healthy template for accurate segmentation of intracranial hemorrhage. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11766, pp. 848–855. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32248-9_94
Liao, R., et al.: An artificial agent for robust image registration. In: Thirty-First AAAI Conference on Artificial Intelligence (2017)
Lim, N.H., Fowkes, M.M.: Radiopaque compound containing diiodotyrosine, 5 Jun 2019, EU Patent EP3490614A1
Ma, K., et al.: Multimodal image registration with deep context reinforcement learning. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017. LNCS, vol. 10433, pp. 240–248. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66182-7_28
Ou, Y., Sotiras, A., Paragios, N., Davatzikos, C.: DRAMMS: deformable registration via attribute matching and mutual-saliency weighting. Med. Image Anal. 15(4), 622–639 (2011)
Papież, B.W., Szmul, A., Grau, V., Brady, J.M., Schnabel, J.A.: Non-local graph-based regularization for deformable image registration. In: Müller, H., et al. (eds.) MICCAI 2019. LNCS, vol. 11766, pp. 199–207. Springer, Cham (2016). https://doi.org/10.1007/978-3-030-32248-9_94
Salehi, S.S.M., Khan, S., Erdogmus, D., Gholipour, A.: Real-time deep pose estimation with geodesic loss for image-to-template rigid registration. IEEE TMI 38(2), 470–481 (2018)
Schnabel, J.A., Heinrich, M.P., Papież, B.W., Brady, J.M.: Advances and challenges in deformable image registration: from image fusion to complex motion modelling. Med. Image Anal. 33, 145–148 (2016)
Kwon, D., et al.: Siamese U-net with healthy template for accurate segmentation of intracranial hemorrhage. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11766, pp. 848–855. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32248-9_94
Sloan, J.M., Goatman, K.A., Siebert, J.P.: Learning rigid image registration-utilizing convolutional neural networks for medical image registration. In: 11th International Joint Conference on Biomedical Engineering Systems and Technologies, pp. 89–99. SCITEPRESS-Science and Technology Publications (2018)
de Vos, B.D., Berendsen, F.F., Viergever, M.A., Sokooti, H., Staring, M., Išgum, I.: A deep learning framework for unsupervised affine and deformable image registration. Med. Image Anal. 52, 128–143 (2019)
Wang, C., Papanastasiou, G., Chartsias, A., Jacenkow, G., Tsaftaris, S.A., Zhang, H.: FIRE: unsupervised bi-directional inter-modality registration using deep networks. arXiv preprint arXiv:1907.05062 (2019)
Wang, X., Girshick, R., Gupta, A., He, K.: Non-local neural networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 7794–7803 (2018)
Zhou, Y., Barnes, C., Lu, J., Yang, J., Li, H.: On the continuity of rotation representations in neural networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 5745–5753 (2019)
Acknowledgement
This work was supported by a Kennedy Trust for Rheumatology Research Studentship, the Centre for OA Pathogenesis Versus Arthritits (Versus Arthritis grant 21621). The authors acknowledge Patricia das Neves Borges as the researcher who collected the preclinical CT dataset, as part of the National Centre for Replacement, Refinement and Reduction of Animals in Research (NC3R grant NC/M000141/1). B. W. Papież acknowledges Rutherford Fund at Health Data Research UK
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Zheng, JQ., Lim, N.H., Papież, B.W. (2020). D-net: Siamese Based Network for Arbitrarily Oriented Volume Alignment. In: Reuter, M., Wachinger, C., Lombaert, H., Paniagua, B., Goksel, O., Rekik, I. (eds) Shape in Medical Imaging. ShapeMI 2020. Lecture Notes in Computer Science(), vol 12474. Springer, Cham. https://doi.org/10.1007/978-3-030-61056-2_6
Download citation
DOI: https://doi.org/10.1007/978-3-030-61056-2_6
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-61055-5
Online ISBN: 978-3-030-61056-2
eBook Packages: Computer ScienceComputer Science (R0)