Abstract
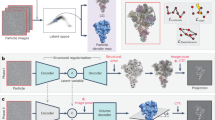
We propose a deep-learning-based reconstruction method for cryo-electron microscopy (Cryo-EM) that can model multiple conformations of a nonrigid biomolecule in a standalone manner. Cryo-EM produces many noisy projections from separate instances of the same but randomly oriented biomolecule. Current methods rely on pose and conformation estimation which are inefficient for the reconstruction of continuous conformations that carry valuable information. We introduce Multi-CryoGAN, which sidesteps the additional processing by casting the volume reconstruction into the distribution matching problem. By introducing a manifold mapping module, Multi-CryoGAN can learn continuous structural heterogeneity without pose estimation nor clustering. We also give a theoretical guarantee of recovery of the true conformations. Our method can successfully reconstruct 3D protein complexes on synthetic 2D Cryo-EM datasets for both continuous and discrete structural variability scenarios. Multi-CryoGAN is the first model that can reconstruct continuous conformations of a biomolecule from Cryo-EM images in a fully unsupervised and end-to-end manner.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Andén, J., Katsevich, E., Singer, A.: Covariance estimation using conjugate gradient for 3D classification in cryo-EM. In: 12th International Symposium on Biomedical Imaging (ISBI), pp. 200–204. IEEE (2015)
Arjovsky, M., Chintala, S., Bottou, L.: Wasserstein generative adversarial networks. In: International Conference on Machine Learning, pp. 214–223 (2017)
Bendory, T., Bartesaghi, A., Singer, A.: Single-particle cryo-electron microscopy: mathematical theory, computational challenges, and opportunities. IEEE Signal Process. Mag. 37(2), 58–76 (2020)
Bora, A., Price, E., Dimakis, A.G.: AmbientGAN: generative models from lossy measurements. In: International Conference on Learning Representations, vol. 2, pp. 5–15 (2018)
Dashti, A., et al.: Trajectories of the ribosome as a Brownian nanomachine. Proc. Natl. Acad. Sci. 111(49), 17492–17497 (2014)
Frank, J., Ourmazd, A.: Continuous changes in structure mapped by manifold embedding of single-particle data in Cryo-EM. Methods 100, 61–67 (2016)
Gadelha, M., Maji, S., Wang, R.: 3D shape induction from 2D views of multiple objects. In: 2017 International Conference on 3D Vision (3DV), pp. 402–411 (2017)
Goodfellow, I., et al.: Generative adversarial nets. In: Advances in Neural Information Processing Systems, pp. 2672–2680 (2014)
Gulrajani, I., Ahmed, F., Arjovsky, M., Dumoulin, V., Courville, A.C.: Improved training of Wasserstein GANs. In: Advances in Neural Information Processing Systems, pp. 5767–5777 (2017)
Gupta, H., McCann, M.T., Donati, L., Unser, M.: CryoGAN: a new reconstruction paradigm for single-particle Cryo-EM via deep adversarial learning. BioRxiv (2020)
Hornik, K., Stinchcombe, M., White, H.: Multilayer feedforward networks are universal approximators. Neural Netw. 2(5), 359–366 (1989)
Karras, T., Aila, T., Laine, S., Lehtinen, J.: Progressive growing of GANs for improved quality, stability, and variation. arXiv:1710.10196 (2017)
Karras, T., Laine, S., Aila, T.: A style-based generator architecture for generative adversarial networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 4401–4410 (2019)
Lederman, R.R., Andén, J., Singer, A.: Hyper-molecules: on the representation and recovery of dynamical structures for applications in flexible macro-molecules in cryo-EM. Inverse Prob. 36(4), 044005 (2020)
Miolane, N., Poitevin, F., Li, Y.T., Holmes, S.: Estimation of orientation and camera parameters from cryo-electron microscopy images with variational autoencoders and generative adversarial networks. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, pp. 970–971 (2020)
Moscovich, A., Halevi, A., Andén, J., Singer, A.: Cryo-EM reconstruction of continuous heterogeneity by Laplacian spectral volumes. Inverse Prob. 36(2), 024003 (2020)
Ourmazd, A.: Cryo-EM, XFELs and the structure conundrum in structural biology. Nat. Methods 16(10), 941–944 (2019)
Punjani, A., Rubinstein, J.L., Fleet, D.J., Brubaker, M.A.: cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14(3), 290–296 (2017)
Scheres, S.H.: RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180(3), 519–530 (2012)
Seitz, E., Acosta-Reyes, F., Schwander, P., Frank, J.: Simulation of cryo-EM ensembles from atomic models of molecules exhibiting continuous conformations. BioRxiv p. 864116 (2019)
Singer, A., Sigworth, F.J.: Computational methods for single-particle electron cryomicroscopy. Ann. Rev. Biomed. Data Sci. 3 (2020)
Sorzano, C.O.S., et al.: Survey of the analysis of continuous conformational variability of biological macromolecules by electron microscopy. Acta Crystallogr. Sect. F Struct. Biol. Commun. 75(1), 19–32 (2019)
Tewari, A., et al.: State of the art on neural rendering. arXiv preprint arXiv:2004.03805 (2020)
Tulsiani, S., Efros, A.A., Malik, J.: Multi-view consistency as supervisory signal for learning shape and pose prediction. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2897–2905 (2018)
Villani, C.: Optimal Transport: Old and New, vol. 338. Springer, Heidelberg (2008)
Zhong, E.D., Bepler, T., Berger, B., Davis, J.H.: CryoDRGN: reconstruction of heterogeneous structures from cryo-electron micrographs using neural networks. bioRxiv (2020)
Acknowledgments
This work is funded by H2020-ERC Grant 692726 (GlobalBioIm).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
1 Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Gupta, H., Phan, T.H., Yoo, J., Unser, M. (2020). Multi-CryoGAN: Reconstruction of Continuous Conformations in Cryo-EM Using Generative Adversarial Networks. In: Bartoli, A., Fusiello, A. (eds) Computer Vision – ECCV 2020 Workshops. ECCV 2020. Lecture Notes in Computer Science(), vol 12535. Springer, Cham. https://doi.org/10.1007/978-3-030-66415-2_28
Download citation
DOI: https://doi.org/10.1007/978-3-030-66415-2_28
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-66414-5
Online ISBN: 978-3-030-66415-2
eBook Packages: Computer ScienceComputer Science (R0)