Abstract
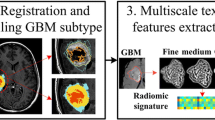
Radiomics based multivariate models are potentially valuable prognostic tool in IDH phenotyping in high-grade gliomas. Radiomics generally involves a set of the histogram and standard texture features based on co-occurrence matrix, run-length matrix, size zone matrix, gray tone matrix, and gray level dependence matrix as described by the Imaging Biomarker Standardization Initiative (IBSI). In this work, we introduce 3D local binary patterns (3D LBP) and local ternary patterns (3D LTP) based histogram features in addition to standard radiomics for capturing subtle phenotypic differences to characterize IDH genotype in high-grade gliomas. These textures are rotationally invariant and are robust to image noise as well as can capture the underlying local differences in the tissue architecture. On a dataset of 64 patients with high-grade glioma scanned at a single institution, we illustrate that LBP and LTP features perform at par with radiomics individually and when combined, could facilitate highest testing accuracy of 84.62% (AUROC: 0.78, sensitivity: 0.83 specificity: 0.86, f1-score: 0.83) using random forest classification. Clinical explainability is achieved via feature ranking and selection where the top 5 features are based on LBP and LTP histogram. Overall, our results illustrate that 3D LBP and LTP histogram features are crucial in creating tumor phenotypic signatures and could be included in the standard radiomics pipeline.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Wank, M., et al.: Human glioma migration and infiltration properties as a target for personalized radiation medicine. Cancers 10(11), 456 (2018)
Louis, D.N., et al.: The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 131(6), 803–820 (2016)
Houillier, C., et al.: IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology 75(17), 1560–1566 (2010)
Jakola, A., et al.: Quantitative texture analysis in the prediction of IDH status in low-grade gliomas. J. Clin. Neurol. Neurosurg. 164, 114–120 (2017)
Chougule, T., Shinde, S., Santosh, V., Saini, J., Ingalhalikar, M.: On Validating Multimodal MRI Based Stratification of IDH Genotype in High Grade Gliomas Using CNNs and Its Comparison to Radiomics. In: Mohy-ud-Din, H., Rathore, S. (eds.) RNO-AI 2019. LNCS, vol. 11991, pp. 53–60. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-40124-5_6
Gore, S., et al.: A review of radiomics and deep predictive modeling in glioma characterization. Academic Radiology, epub ahead of print (2020). https://doi.org/10.1010/j.acra.2020.06.016
Zhao, G., et al.: Dynamic texture recognition using local binary patterns with an application to facial expressions. IEEE Trans. Pattern Anal. Mach. Intell. 29(6), 915–928 (2007)
Ahonen, T., et al.: Face description with local binary patterns: application to face recognition. IEEE Trans. Pattern Anal. Mach. Intell. 28(12), 2037–2041 (2006)
Samal, A., et al.: Texture as the basis for individual tree identification. Inf. Sci. 176, 565–576 (2006)
Avants, B.B., et al.: Advanced normalization tools (ANTS), Insight j 2.365, pp. 1–35 (2009)
Smith, S.M.: Fast robust automated brain extraction. Hum. Brain Mapp. 17(3), 143–155 (2002)
Kamnitsas, K., et al.: DeepMedic for brain tumor segmentation. In: Crimi, A., Menze, B., Maier, O., Reyes, M., Winzeck, S., Handels, H. (eds.) BrainLes 2016. LNCS, vol. 10154, pp. 138–149. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-55524-9_14
Menze, B.H., et al.: The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging 34(10), 1993–2024 (2015). https://doi.org/10.1109/TMI.2014.2377694
Bakas, S., et al.: Advancing the Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Nature Sci. Data 4, 170117 (2017). https://doi.org/10.1038/sdata.2017.117
Bakas, S., et al.: Identifying the Best Machine Learning Algorithms for Brain Tumor Segmentation, Progression Assessment, and Overall Survival Prediction in the BRATS Challenge, arXiv preprint arXiv:1811.02629 (2018)
Ojala, T., et al.: A comparative study of texture measures with classification based on feature distributions. Pattern Recogn. 29(1), 51–59 (1996)
Ojala, T., et al.: Multiresolution gray-scale and rotation invariant texture classification with Local Binary Patterns. IEEE Trans. Pattern Anal. Mach. Intell. 24(7), 971–987 (2002)
Tan, X., et al.: Enhanced local texture feature sets for face recognition under difficult lighting conditions. IEEE Trans. Image Process. 19(6), 1635–1650 (2010)
Griethuysen, J.J.M., et al.: Computational radiomics system to decode the radiographic phenotype. Can. Res. 77(21), e104–e107 (2017)
Pedregosa et al.: Scikit-learn: Machine Learning in Python. JMLR 12, 2825–2830 (2011)
Darst, B.F., et al.: Using recursive feature elimination in random forest to account for correlated variables in high dimensional data. BMC Genet. 19, 65 (2018)
Chaddad, A., et al.: Predicting the gene status and survival outcome of lower grade glioma patients with multimodal MRI features. IEEE Access 7, 75976–75984 (2019). https://doi.org/10.1109/ACCESS.2019.2920396
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Gore, S., Chougule, T., Saini, J., Ingalhalikar, M., Jagtap, J. (2020). Local Binary and Ternary Patterns Based Quantitative Texture Analysis for Assessment of IDH Genotype in Gliomas on Multi-modal MRI. In: Kia, S.M., et al. Machine Learning in Clinical Neuroimaging and Radiogenomics in Neuro-oncology. MLCN RNO-AI 2020 2020. Lecture Notes in Computer Science(), vol 12449. Springer, Cham. https://doi.org/10.1007/978-3-030-66843-3_23
Download citation
DOI: https://doi.org/10.1007/978-3-030-66843-3_23
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-66842-6
Online ISBN: 978-3-030-66843-3
eBook Packages: Computer ScienceComputer Science (R0)