Abstract
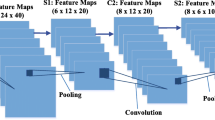
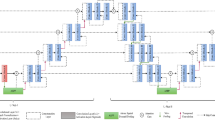
The rise of deep learning techniques, such as a convolutional neural network (CNN) in solving medical image problems, offered fascinating results that motivated researchers to design automatic diagnostic systems. Image segmentation is one of the crucial and challenging steps in the design of a computer-aided diagnosis system owing to the presence of low contrast between skin lesion and background, noise artifacts, color variations, and irregular lesion boundaries. In this paper, we propose a modified and improved encoder-decoder architecture with a smaller network depth and a smaller number of kernels to enhance the segmentation process. The network performs segmentation for skin cancer images to obtain information about the infected area. The proposed model utilizes the power of the VGG19 network’s weight layers for calculating rich features. The deconvolutional layers were designed to regain spatial information of the image. In addition to this, optimized training parameters were adopted to further improve the network’s performance. The designed network was evaluated for two publicly available benchmarked datasets ISIC, and PH2 consists of dermoscopic skin cancer images. The experimental observations proved that the proposed network achieved the higher average values of segmentation accuracy 95.67%, IoU 96.70%, and BF-score of 89.20% on ISIC 2017 and accuracy 98.50%, IoU 93.25%, and BF-score 84.08% on PH2 datasets as compared to other state-of-the-art algorithms on the same datasets.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Skourt, B.A., El Hassani, A., Majda, A.: Lung CT image segmentation using deep neural networks. Procedia Comput. Sci. 127, 109–113 (2018)
Zhang, Y., et al.: Automatic breast and fibroglandular tissue segmentation in breast MRI using deep learning by a fully-convolutional residual neural network U-net. Acad. Radiol. 26(11), 1526–1535 (2019)
Yu, S., Xiao, D., Frost, S., Kanagasingam, Y.: Robust optic disc and cup segmentation with deep learning for glaucoma detection. Comput. Med. Imaging Graph. 74, 61–71 (2019)
Wu, D., Xu, L., Zhang, R., Zhang, H., Ren, L., Zhang, Y.-T.: Continuous cuff-less blood pressure estimation based on combined information using deep learning approach. J. Med. Imaging Health Inform. 8(6), 1290–1299 (2018)
Skin Cancer-Index (2018). https://www.isic-archive.com. Accessed 2019
What is Melanoma Skin Cancer? https://www.cancer.org/cancer/melanoma-skin-cancer/about/what-is-melanoma.html. Accessed 2019
Bogo, F., Peruch, F., Fortina, A.B., Peserico, E.: Where’s the lesion?: Variability in human and automated segmentation of dermoscopy images of melanocytic skin lesions. In: Dermoscopy Image Analysis, pp. 82–110. CRC Press (2015)
Types of Skin Cancer. https://www.everydayhealth.com/skin-cancer/types. Accessed 2020
Sridevi, M., Mala, C.: A survey on monochrome image segmentation methods. Procedia Technol. 6, 548–555 (2012)
Barcelos, C.A.Z., Pires, V.: An automatic based nonlinear diffusion equations scheme for skin lesion segmentation. Appl. Math. Comput. 215(1), 251–261 (2009)
de Souza Ganzeli, H., Bottesini, J.G., de Oliveira Paz, L., Ribeiro, M.F.S.: Skan: Skin scanner-system for skin cancer detection using adaptive techniques. IEEE Latin Am. Trans. 9(2), 206–212 (2011)
Norton, K.A., et al.: Three-phase general border detection method for dermoscopy images using non-uniform illumination correction. Skin Res. Technol. 18(3), 290–300 (2012)
Qaisar Abbas, M., Celebi, E., Fondón, I.: Computer-aided pattern classification system for dermoscopy images. Skin Res. Technol. 18(3), 278–289 (2012)
Garnavi, R., Aldeen, M., Celebi, M.E., Varigos, G., Finch, S.: Border detection in dermoscopy images using hybrid thresholding on optimized color channels. Comput. Med. Imaging Graph. 35(2), 105–115 (2011)
Ma, Z., Tavares, J.M.R.: A novel approach to segment skin lesions in dermoscopic images based on a deformable model. IEEE J. Biomed. Health Inform. 20(2), 615–623 (2015)
Cavalcanti, P.G., Scharcanski, J.: A coarse-to-fine approach for segmenting melanocytic skin lesions in standard camera images. Comput. Methods Programs Biomed. 112(3), 684–693 (2013)
Shan, P.: Image segmentation method based on K-mean algorithm. EURASIP J. Image Video Process. 2018(1), 1–9 (2018). https://doi.org/10.1186/s13640-018-0322-6
Choudhry, M.S., Kapoor, R.: Performance analysis of fuzzy C-means clustering methods for MRI image segmentation. Procedia Comput. Sci. 89, 749–758 (2016)
Kasmi, R., Mokrani, K.: Classification of malignant melanoma and benign skin lesions: implementation of automatic ABCD rule. IET Image Proc. 10(6), 448–455 (2016)
Vasconcelos, F., Medeiros, A., Peixoto, S., Filho, P.: Automatic skin lesions segmentation based on a new morphological approach via geodesic active contour. Cognit. Syst. Res. 55, 44–59 (2019)
Bayraktar, M., Kockara, S., Halic, T., Mete, M., Wong, H.K., Iqbal, K.: Local edge-enhanced active contour for accurate skin lesion border detection. BMC Bioinform. 20(2), 91 (2019). https://doi.org/10.1186/s12859-019-2625-8
Manjón, J.V., et al.: MRI white matter lesion segmentation using an ensemble of neural networks and overcomplete patch-based voting. Comput. Med. Imaging Graph. 69, 43–51 (2018)
Tan, T.Y., Zhang, L., Lim, C.P., Fielding, B., Yu, Y., Anderson, E.: Evolving ensemble models for image segmentation using enhanced particle swarm optimization. IEEE Access 7, 34004–34019 (2019)
Alshayeji, M.H., Al-Rousan, M.A., Ellethy, H., Abed, S.: An efficient multiple sclerosis segmentation and detection system using neural networks. Comput. Electr. Eng. 71, 191–205 (2018)
Cernazanu-Glavan, C., Holban, S.: Segmentation of bone structure in X-ray images using convolutional neural network. Adv. Electr. Comput. Eng 13(1), 87–94 (2013)
Melinščak, M., Prentašić, P., Lončarić, S.: Retinal vessel segmentation using deep neural networks. In: 10th International Conference on Computer Vision Theory and Applications (VISAPP 2015) (2015)
Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 3431–3440 (2015)
Hong, S., Noh, H., Han, B.: Decoupled deep neural network for semi-supervised semantic segmentation. In: Advances in Neural Information Processing Systems, pp. 1495–1503 (2015)
Badrinarayanan, V., Kendall, A., Cipolla, R.: Segnet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 39(12), 2481–2495 (2017)
Ronneberger, O., Fischer, P., Brox, T.: U-net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Okur, E., Turkan, M.: A survey on automated melanoma detection. Eng. Appl. Artif. Intell. 73, 50–67 (2018)
Hesamian, M.H., Jia, W., He, X., Kennedy, P.: Deep learning techniques for medical image segmentation: achievements and challenges. J. Digit. Imaging 32(4), 582–596 (2019). https://doi.org/10.1007/s10278-019-00227-x
Yang, X., Zeng, Z., Yeo, S.Y., Tan, C., Tey, H.L., Su, Y.: A novel multi-task deep learning model for skin lesion segmentation and classification. arXiv preprint arXiv:1703.01025 (2017)
Kawahara, J., BenTaieb, A., Hamarneh, G.: Deep features to classify skin lesions. In: 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), pp. 1397–1400. IEEE (2016)
Nida, N., Irtaza, A., Javed, A., Yousaf, M.H., Mahmood, M.T.: Melanoma lesion detection and segmentation using deep region based convolutional neural network and fuzzy C-means clustering. Int. J. Med. Inform. 124, 37–48 (2019)
Nasr-Esfahani, E., et al.: Dense fully convolutional network for skin lesion segmentation arXiv preprint arXiv:1712.10207 (2017)
Vesal, S., Ravikumar, N., Maier, A.: SkinNet: a deep learning framework for skin lesion segmentation arXiv preprint arXiv:1806.09522 (2018)
Zhang, X.: Melanoma segmentation based on deep learning. Comput. Assist. Surg. 22(s1), 267–277 (2017)
Yu, L., Chen, H., Dou, Q., Qin, J., Heng, P.-A.: Automated melanoma recognition in dermoscopy images via very deep residual networks. IEEE Trans. Med. Imaging 36(4), 994–1004 (2016)
Simonyan, K., Zisserman, A.: Very deep convolutional networks for large-scale image recognition arXiv preprint arXiv:1409.1556 (2014)
Bishop, C.M.: Pattern Recognition and Machine Learning. Springer, New York (2006)
The International Skin Imaging Collaboration. www.isic-archive.com. Accessed 2019
Dermofit Image Library. https://licensing.edinburgh-innovations.ed.ac.uk/. Accessed 2019
Ninh, Q.C., Tran, T.-T., Tran, T.T., Tran, T.A.X., Pham, V.-T.: Skin lesion segmentation based on modification of SegNet neural networks. In: 2019 6th NAFOSTED Conference on Information and Computer Science (NICS), pp. 575–578. IEEE (2019)
Jahanifar, M., Tajeddin, N.Z., Asl, B.M., Gooya, A.: Supervised saliency map driven segmentation of lesions in dermoscopic images. IEEE J. Biomed. Health Inform. 23(2), 509–518 (2018)
Yuan, Y., Chao, M., Lo, Y.-C.: Automatic skin lesion segmentation using deep fully convolutional networks with Jaccard distance. IEEE Trans. Med. Imaging 36(9), 1876–1886 (2017)
Bi, L., Kim, J., Ahn, E., Kumar, A., Feng, D., Fulham, M.: Step-wise integration of deep class-specific learning for dermoscopic image segmentation. Pattern Recogn. 85, 78–89 (2019)
Bi, L., Kim, J., Ahn, E., Kumar, A., Fulham, M., Feng, D.: Dermoscopic image segmentation via multi-stage fully convolutional networks. IEEE Trans. Biomed. Eng. 64, 2065–2074 (2017)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Kaur, R., GholamHosseini, H., Sinha, R. (2021). Deep Learning in Medical Applications: Lesion Segmentation in Skin Cancer Images Using Modified and Improved Encoder-Decoder Architecture. In: Nguyen, M., Yan, W.Q., Ho, H. (eds) Geometry and Vision. ISGV 2021. Communications in Computer and Information Science, vol 1386. Springer, Cham. https://doi.org/10.1007/978-3-030-72073-5_4
Download citation
DOI: https://doi.org/10.1007/978-3-030-72073-5_4
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-72072-8
Online ISBN: 978-3-030-72073-5
eBook Packages: Computer ScienceComputer Science (R0)