Abstract
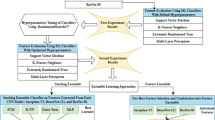
Gliomas are brain tumours with a high mortality rate. There are various grades and sub-types of this tumour, and the treatment procedure varies accordingly. Clinicians and oncologists diagnose and categorise these tumours based on visual inspection of radiology and histology data. However, this process can be time-consuming and subjective. The computer-assisted methods can help clinicians to make better and faster decisions. In this paper, we propose a pipeline for automatic classification of gliomas into three sub-types: oligodendroglioma, astrocytoma, and glioblastoma, using both radiology and histopathology images. The proposed approach implements distinct classification models for radiographic and histologic modalities and combines them through an ensemble method. The classification algorithm initially carries out tile-level (for histology) and slice-level (for radiology) classification via a deep learning method, then tile/slice-level latent features are combined for a whole-slide and whole-volume sub-type prediction. The classification algorithm was evaluated using the data set provided in the CPM-RadPath 2020 challenge. The proposed pipeline achieved the F1-Score of 0.886, Cohen’s Kappa score of 0.811 and Balance accuracy of 0.860. The ability of the proposed model for end-to-end learning of diverse features enables it to give a comparable prediction of glioma tumour sub-types.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Abdalla, G., et al.: The diagnostic role of diffusional kurtosis imaging in glioma grading and differentiation of gliomas from other intra-axial brain tumours: a systematic review with critical appraisal and meta-analysis. Neuroradiology 62(7), 791–802 (2020). https://doi.org/10.1007/s00234-020-02425-9
Bertero, L., Cassoni, P.: Classification of tumours of the central nervous system. In: Bartolo, M., Soffietti, R., Klein, M. (eds.) Neurorehabilitation in Neuro-Oncology, pp. 21–36. Springer, Cham (2019). https://doi.org/10.1007/978-3-319-95684-8_3
Campanella, G., et al.: Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 25(8), 1301–1309 (2019)
Chan, H.-W., Weng, Y.-T., Huang, T.-Y.: Automatic classification of brain tumor types with the MRI scans and histopathology images. In: Crimi, A., Bakas, S. (eds.) BrainLes 2019. LNCS, vol. 11993, pp. 353–359. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-46643-5_35
Decuyper, M., Bonte, S., Deblaere, K., Van Holen, R.: Automated MRI based pipeline for glioma segmentation and prediction of grade, IDH mutation and 1p19q co-deletion. arXiv preprint arXiv:2005.11965 (2020)
Dhermain, F.G., Hau, P., Lanfermann, H., Jacobs, A.H., van den Bent, M.J.: Advanced MRI and PET imaging for assessment of treatment response in patients with gliomas. Lancet Neurol. 9(9), 906–920 (2010)
Hamidinekoo, A., Denton, E., Rampun, A., Honnor, K., Zwiggelaar, R.: Deep learning in mammography and breast histology, an overview and future trends. Med. Image Anal. 47, 45–67 (2018)
Huang, G., Liu, Z., Van Der Maaten, L., Weinberger, K.Q.: Densely connected convolutional networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 4700–4708 (2017)
Jameson, J.L., Longo, D.L.: Precision medicine-personalized, problematic, and promising. Obstet. Gynecol. Surv. 70(10), 612–614 (2015)
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980 (2014)
Kumar, V., Abbas, A.K., Aster, J.C.: Robbins Basic Pathology, 10th edn. Elsevier, Philadelphia (2018)
Kurc, T., et al.: Segmentation and classification in digital pathology for glioma research: challenges and deep learning approaches. Front. Neurosci. 14, 27 (2020)
Ma, X., Jia, F.: Brain tumor classification with multimodal MR and pathology images. In: Crimi, A., Bakas, S. (eds.) BrainLes 2019. LNCS, vol. 11993, pp. 343–352. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-46643-5_34
Pei, L., Vidyaratne, L., Hsu, W.-W., Rahman, M.M., Iftekharuddin, K.M.: Brain tumor classification using 3D convolutional neural network. In: Crimi, A., Bakas, S. (eds.) BrainLes 2019. LNCS, vol. 11993, pp. 335–342. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-46643-5_33
Wang, X., et al.: Weakly supervised deep learning for whole slide lung cancer image analysis. IEEE Trans. Cybern. 50(9), 3950–3962 (2019)
Xue, Y., et al.: Brain tumor classification with tumor segmentations and a dual path residual convolutional neural network from MRI and pathology images. In: Crimi, A., Bakas, S. (eds.) BrainLes 2019. LNCS, vol. 11993, pp. 360–367. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-46643-5_36
Ye, Z., et al.: Diffusion histology imaging combining diffusion basis spectrum imaging (DBSI) and machine learning improves detection and classification of glioblastoma pathology. Clin. Cancer Res. (2020). https://doi.org/10.1158/1078-0432.CCR-20-0736
Acknowledgements
Azam Hamidinekoo acknowledges the support by Children’s Cancer and Leukaemia Group (CCLGA201906). Tomasz Pieciak acknowledges the Polish National Agency for Academic Exchange for project PN/BEK/2019/1/00421 under the Bekker programme. Maryam Afzali was supported by a Wellcome Trust Investigator Award (096646/Z/11/Z). Otar Akanyeti acknowledges Sér Cymru Cofund II Research Fellowship grant. Yinyin Yuan acknowledges funding from Cancer Research UK Career Establishment Award (CRUK C45982/A21808), CRUK Early Detection Program Award (C9203/A28770), CRUK Sarcoma Accelerator (C56167/A29363), CRUK Brain Tumour Award (C25858/A28592), Rosetrees Trust (A2714), Children’s Cancer and Leukaemia Group (CCLGA201906), NIH U54 CA217376, NIH R01 CA185138, CDMRP BreastCancer Research Program Award BC132057, European Commission ITN (H2020-MSCA-ITN-2019), and The Royal Marsden/ICR National Institute of Health Research Biomedical Research Centre. This research was also supported in part by PLGrid Infrastructure. Tomasz Pieciak acknowledges Mr. Maciej Czuchry from CYFRONET Academic Computer Centre (Kraków, Poland) for outstanding support.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Hamidinekoo, A., Pieciak, T., Afzali, M., Akanyeti, O., Yuan, Y. (2021). Glioma Classification Using Multimodal Radiology and Histology Data. In: Crimi, A., Bakas, S. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2020. Lecture Notes in Computer Science(), vol 12659. Springer, Cham. https://doi.org/10.1007/978-3-030-72087-2_45
Download citation
DOI: https://doi.org/10.1007/978-3-030-72087-2_45
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-72086-5
Online ISBN: 978-3-030-72087-2
eBook Packages: Computer ScienceComputer Science (R0)