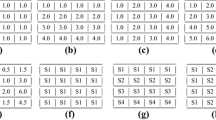
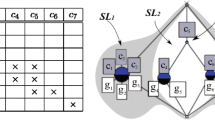
Abstract
Formal Concept Analysis has been widely applied to identify differently expressed genes among microarray data. Top-K Formal Concepts are identified as efficient in generating most important Formal Concepts. To the best of our knowledge, no currently available algorithm is able to perform this challenging task. Therefore, we introduce Top-BicMiner, a new method for mining biclusters from gene expression data through Top-k Formal Concepts. It performs the extraction of the sets of both positive and negative correlations biclusters. Top-BicMiner relies on Formal concept analysis as well as a specific discretization method. Extensive experiments, carried out on real-life datasets, shed light on Top-BicMiner’s ability to identify statistically and biologically significant biclusters.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Notes
- 1.
The extraction of the formal concepts is carried out through the invocation of the efficient LCM algorithm [23].
- 2.
In fact, coherent formal concepts having an intersection size above or equal to the given threshold \(\alpha 1\) belong to the same bicluster, while those with an intersection value below it, do not.
.
- 3.
Available at https://github.com/mehdi-kaytoue/trimax.
- 4.
Available at http://arep.med.harvard.edu/biclustering/.
- 5.
Available at http://arep.med.harvard.edu/biclustering/.
- 6.
Available at http://llama.mshri.on.ca/funcassociate/.
- 7.
The best biclusters have an adjusted p-value less than 0.001%.
- 8.
Available at https://www.yeastgenome.org/goTermFinder.
- 9.
References
Ayadi, W., Elloumi, M., Hao, J.K.: A biclustering algorithm based on a bicluster enumeration tree: application to DNA microarray data. BioData Mining 2, 9 (2009)
Ayadi, W., Elloumi, M., Hao, J.K.: Bicfinder: a biclustering algorithm for microarray data analysis. Knowl. Inf. Syst. 30(2), 341–358 (2012)
Ayadi, W., Hao, J.: A memetic algorithm for discovering negative correlation biclusters of DNA microarray data. Neurocomputing 145, 14–22 (2014). https://doi.org/10.1016/j.neucom.2014.05.074
Ben-Dor, A., Chor, B., Karp, R.M., Yakhini, Z.: Discovering local structure in gene expression data: the order-preserving submatrix problem. J. Comput. Biol. 10(3/4), 373–384 (2003)
Bergmann, S., Ihmels, J., Barkai, N.: Defining transcription modules using large-scale gene expression data. Bioinformatics 20(13), 1993–2003 (2004)
Madeira, S.C., Oliveira, A.L.: Biclustering algorithms for biological data analysis: a survey. IEEE Trans. Comput. Biol. Bioinf. 1, 24–45 (2004)
Cheng, Y., Church, G.M.: Biclustering of expression data. In: Proceedings of ISMB, UC San Diego, California, pp. 93–103 (2000)
Ganter, B., Wille, R.: Formal Concept Analysis - Mathematical Foundations. Springer, Heidelberg (1999). https://doi.org/10.1007/978-3-642-59830-2
Henriques, R., Antunes, C., Madeira, S.C.: Methods for the efficient discovery of large item-indexable sequential patterns. In: New Frontiers in Mining Complex Patterns - Second International Workshop, NFMCP 2013, Held in Conjunction with ECML-PKDD 2013, Prague, Czech Republic, 27 September 2013, Revised Selected Papers, pp. 100–116 (2013). https://doi.org/10.1007/978-3-319-08407-7_7
Henriques, R., Madeira, S.C.: Bicspam: flexible biclustering using sequential patterns. BMC Bioinf. 15, 130 (2014). https://doi.org/10.1186/1471-2105-15-130
Henriques, R., Madeira, S.C.: Bic2pam: constraint-guided biclustering for biological data analysis with domain knowledge. Algorithms Molec. Biol. 11, 23 (2016). https://doi.org/10.1186/s13015-016-0085-5
Houari, A., Ayadi, W., Yahia, S.B.: Discovering low overlapping biclusters in gene expression data through generic association rules. In: Bellatreche, L., Manolopoulos, Y. (eds.) MEDI 2015. LNCS, vol. 9344, pp. 139–153. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-23781-7_12
Houari, A., Ayadi, W., Yahia, S.B.: Mining negative correlation biclusters from gene expression data using generic association rules. In: Zanni-Merk, C., Frydman, C.S., Toro, C., Hicks, Y., Howlett, R.J., Jain, L.C. (eds.) Knowledge-Based and Intelligent Information & Engineering Systems: Proceedings of the 21st International Conference KES-2017, Marseille, France, 6–8 September 2017, Procedia Computer Science, vol. 112, pp. 278–287. Elsevier (2017). https://doi.org/10.1016/j.procs.2017.08.262
Houari, A., Ayadi, W., Yahia, S.B.: NBF: an fca-based algorithm to identify negative correlation biclusters of DNA microarray data. In: Barolli, L., Takizawa, M., Enokido, T., Ogiela, M.R., Ogiela, L., Javaid, N. (eds.) 32nd IEEE International Conference on Advanced Information Networking and Applications, AINA 2018, Krakow, Poland, 16–18 May 2018, pp. 1003–1010. IEEE Computer Society (2018). https://doi.org/10.1109/AINA.2018.00146
Houari, A., Ayadi, W., Yahia, S.B.: A new fca-based method for identifying biclusters in gene expression data. Int. J. Mach. Learn. Cybern. 9(11), 1879–1893 (2018). https://doi.org/10.1007/s13042-018-0794-9
Hwang, C.L., Yoon, K.: Methods for multiple attribute decision making. In: Multiple Attribute Decision Making, pp. 58–191. Springer, Heidelberg (1981). https://doi.org/10.1007/978-3-642-48318-9_3
Kaytoue, M., Kuznetsov, S.O., Macko, J., Napoli, A.: Biclustering meets triadic concept analysis. Ann. Math. Artif. Intell. 70(1–2), 55–79 (2014). https://doi.org/10.1007/s10472-013-9379-1
Kuznetsov, S.O.: Stability as an estimate of degree of substantiation of hypotheses derived on the basis of operational similarity. Nauchno-Tekhnichekaya Informatisiya Seriya 2-Informatsionnye Protsessy I Sistemy (12), 21–29 (1990)
Li, X., Shao, M.-W., Zhao, X.-M.: Constructing lattice based on irreducible concepts. Int. J. Mach. Learn. Cybern. 8(1), 109–122 (2016). https://doi.org/10.1007/s13042-016-0587-y
Mouakher, A., Ben Yahia, S.: Qualitycover: efficient binary relation coverage guided by induced knowledge quality. Inf. Sci. 355, 58–73 (2016)
Nepomuceno, J.A., Troncoso, A., Aguilar-Ruiz, J.S.: Scatter search-based identification of local patterns with positive and negative correlations in gene expression data. Appl. Soft Comput. 35, 637–651 (2015). https://doi.org/10.1016/j.asoc.2015.06.019
Prelic, A., et al.: A systematic comparison and evaluation of biclustering methods for gene expression data. Bioinformatics 22(9), 1122–1129 (2006)
Uno, T., Asai, T., Uchida, Y., Arimura, H.: An efficient algorithm for enumerating closed patterns in transaction databases. In: Discovery Science, 7th International Conference, DS 2004, Padova, Italy, 2–5 October 2004, Proceedings, pp. 16–31 (2004). https://doi.org/10.1007/978-3-540-30214-8_2
Zanakis, S.H., Solomon, A., Wishart, N., Dublish, S.: Multi-attribute decision making: a simulation comparison of select methods. Eur. J. Oper. Res. 107(3), 507–529 (1998)
Zhao, Y., Yu, J., Wang, G., Chen, L., Wang, B., Yu, G.: Maximal subspace coregulated gene clustering. IEEE Trans. Knowl. Data Eng. 20(1), 83–98 (2008). https://doi.org/10.1109/TKDE.2007.190670
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Houari, A., Ben Yahia, S. (2021). Top-K Formal Concepts for Identifying Positively and Negatively Correlated Biclusters. In: Attiogbé, C., Ben Yahia, S. (eds) Model and Data Engineering. MEDI 2021. Lecture Notes in Computer Science(), vol 12732. Springer, Cham. https://doi.org/10.1007/978-3-030-78428-7_13
Download citation
DOI: https://doi.org/10.1007/978-3-030-78428-7_13
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-78427-0
Online ISBN: 978-3-030-78428-7
eBook Packages: Computer ScienceComputer Science (R0)