Abstract
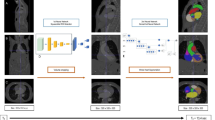
Whole heart segmentation significantly improves the diagnostic value of computed tomography images. Since manual segmentation is very time consuming, efforts have been made to automate this process, especially using deep learning methods such as convolutional neural networks (CNN). This paper considers how preprocessing of computed tomography with the Statistical Dominance Algorithm would affect the results of segmentation obtained with CNNs. Segmentation results were compared to original CTs and processed images. Compared to unprocessed data, improvements in segmentation accuracy were obtained after image processing. Using three-fold cross-validation, the average Dice similarity coefficient that was achieved was 0.811 for unprocessed images and 0.863 for processed images.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Bielecka, M., Obuchowicz, R., Korkosz, M.: The shape language in application to the diagnosis of cervical vertebrae pathology. PLOS ONE 13, e0204,546 (2018). https://doi.org/10.1371/journal.pone.0204546
Chen, C., et al.: Deep learning for cardiac image segmentation: a review. Front. Cardiovasc. Med. 7 (2020). https://doi.org/10.3389/fcvm.2020.00025
Fedorov, A., et al.: 3D slicer as an image computing platform for the quantitative imaging network. Magn. Reson. Imaging 30, 1323–41 (2012). https://doi.org/10.1016/j.mri.2012.05.001
Hesamian, M.H., Jia, W., He, X., Kennedy, P.: Deep learning techniques for medical image segmentation: achievements and challenges. J. Digit. Imaging 32 (2019). https://doi.org/10.1007/s10278-019-00227-x
Kempny, A., Piórkowski, A.: CT2TEE - a novel, internet-based simulator of transoesophageal echocardiography in congenital heart disease. Kardiol. Pol. 68(3), 374–379 (2010)
Kikinis, R., Pieper, S.D., Vosburgh, K.G.: 3D Slicer: a platform for subject-specific image analysis, visualization, and clinical support. pp. 277–289. Springer, New York (2014). https://doi.org/10.1007/978-1-4614-7657-3_19
Li, C., et al.: An 8-layer residual u-net with deep supervision for segmentation of the left ventricle in cardiac CT angiography. Comput. Methods Programs Biomed. 200, 105,876 (2020). https://doi.org/10.1016/j.cmpb.2020.105876
Litjens, G., et al.: State-of-the-art deep learning in cardiovascular image analysis. JACC: Cardiovasc. Imaging 12, 1549–1565 (2019). https://doi.org/10.1016/j.jcmg.2019.06.009
Liu, T., Tian, Y., Zhao, S., Huang, X., Wang, Q.: Automatic whole heart segmentation using a two-stage u-net framework and an adaptive threshold window. IEEE Access 7, 1 (2019). https://doi.org/10.1109/ACCESS.2019.2923318
Lou, Z., Huo, W., Le, K., Tian, X.: Whole heart auto segmentation of cardiac CT images using u-net based GAN, pp. 192–196 (2020). https://doi.org/10.1109/CISP-BMEI51763.2020.9263532
Nurzynska, K.: Deep learning as a tool for automatic segmentation of corneal endothelium images. Symmetry 10(3) (2018). https://doi.org/10.3390/sym10030060
Nurzynska, K., Mikhalkin, A., Piorkowski, A.: CAS: cell annotation software - research on neuronal tissue has never been so transparent. Neuroinformatics 15, 365–382 (2017)
Piorkowski, A.: A statistical dominance algorithm for edge detection and segmentation of medical images. In: Information Technologies in Medicine. Advances in Intelligent Systems and Computing, vol. 471, pp. 3–14. Springer (2016)
Piórkowski, A., Kempny, A.: The transesophageal echocardiography simulator based on computed tomography images. IEEE Trans. Biomed. Eng. 60(2), 292–299 (2013)
Ronneberger, O., Fischer, P., Brox, T.: U-net: convolutional networks for biomedical image segmentation. Lecture Notes in Computer Science, vol. 9351 (2015)
Wang, C., Smedby, O.: Automatic whole heart segmentation using deep learning and shape context, pp. 242–249. Springer (2018). https://doi.org/10.1007/978-3-319-75541-0_26
Wang, W., Ye, C., Zhang, S., Xu, Y., Wang, K.: Improving whole-heart CT image segmentation by attention mechanism. IEEE Access 8, 14579–14587 (2020). https://doi.org/10.1109/ACCESS.2019.2961410
Ye, C., Wang, W., Zhang, S., Wang, K.: Multi-depth fusion network for whole-heart CT image segmentation. IEEE Access PP, 1 (2019). https://doi.org/10.1109/ACCESS.2019.2899635
Yuan, X., Zhu, Y., Wang, Y.: Attention based encoder-decoder network for cardiac semantic segmentation. In: 2020 Chinese Automation Congress (CAC), pp. 4578–4582 (2020). https://doi.org/10.1109/CAC51589.2020.9326844
Zhuang, X.: Challenges and methodologies of fully automatic whole heart segmentation: a review. J. Healthcare Eng. 4, 371–408 (2013). https://doi.org/10.1260/2040-2295.4.3.371
Zhuang, X., et al.: Evaluation of algorithms for multi-modality whole heart segmentation: an open-access grand challenge. Med. Image Anal. 58, 101,537 (2019). https://doi.org/10.1016/j.media.2019.101537
Zhuang, X., Shen, J.: Multi-scale patch and multi-modality atlases for whole heart segmentation of MRI. Med. Image Anal. 31 (2016). https://doi.org/10.1016/j.media.2016.02.006
Acknowledgement
This work was supported by the European Union under the Regional Operational Programme for Malopolska Region 2014–2020, project no: RPMP.01.02.01-12-0027/19.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Lasek, J. (2022). The Impact of Data Preprocessing on the Accuracy of CNN-Based Heart Segmentation. In: Choraś, M., Choraś, R.S., Kurzyński, M., Trajdos, P., Pejaś, J., Hyla, T. (eds) Progress in Image Processing, Pattern Recognition and Communication Systems. CORES IP&C ACS 2021 2021 2021. Lecture Notes in Networks and Systems, vol 255. Springer, Cham. https://doi.org/10.1007/978-3-030-81523-3_17
Download citation
DOI: https://doi.org/10.1007/978-3-030-81523-3_17
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-81522-6
Online ISBN: 978-3-030-81523-3
eBook Packages: Intelligent Technologies and RoboticsIntelligent Technologies and Robotics (R0)